Avicenna Journal of Clinical Microbiology and Infection. 8(1):1-4.
doi: 10.34172/ajcmi.2021.01
Original Article
Plasmid-Mediated Antibiotic-Resistant Pattern of Lactobacillus spp. Isolated From Dairy Products
Fatemeh Shafiei Seifabadi 1, Majid Baserisalehi 1, * 
Author information:
1Department of Microbiology, Islamic Azad University, Kazerun Branch, Kazerun, Iran
*
Corresponding author: Majid Baserisalehi, Department of Microbiology, Islamic Azad University, Kazerun Branch, Kazerun, Iran. Email:
majidbaseri@hotmail.com
Abstract
Background: Microorganisms have potent activity for transferring antibiotic-resistant genes with either chromosomally- or plasmid-mediated characteristics. The purpose of this study was to isolate Lactobacillus from different commercial products and evaluate their potential in antibiotic-resistant development. Chromosomally or plasmid-mediated resistant genes were investigated as well.
Methods: In total, Lactobacillus strains were isolated from 20 commercial dairy product samples such as cheese and yoghurt. The isolates were phenotypic and molecularly identified and their antibiotic-resistant properties were assessed by the disk-diffusion method. Finally, the plasmid-mediated antibiotic resistant characters of the isolates were evaluated by plasmid curing via evaluated temperatures and acridine orange methods.
Results: Five strains Lactobacillus paracasei, L. rhamnosus, L. casei, L. plantarum, and L. fermentum were isolated different products. The results of the antibiotic susceptibility assay indicated that all strains were susceptible to amoxicillin and imipenem and resistant to ciprofloxacin and vancomycin. Furthermore, different responses were observed among the isolates against streptomycin and gentamicin. The evaluation of plasmid-mediated antibiotic resistance in the isolates revealed that streptomycin and gentamicin-resistant characters were of plasmid-mediated type in L. rhamnosus and L. plantarum strains.
Conclusions: In general, our finding demonstrated that some commercial Lactobacillus strains harboured antibiotic-resistant genes. These genes can be located either in chromosome or plasmid group. Hence, the frequency of antibiotic-resistant pathogenic bacteria might be increased after consuming some dairy products because of the horizontal transfer of antibiotic-resistance genes among the bacteria.
Keywords: Lactobacillus, Dairy products, Antibiotics, Plasmid-mediated gene
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Probiotics are living microorganisms with beneficial effects for the host if consumed in sufficient quantities. These microorganisms enhance intestinal health and modulate the immune system (1,2). Due to the increased demand for natural drugs, probiotic bacteria have been added to a number of food products, especially dairy foods (3). Recently, a biological system is available on the market in the form of capsules and tablets.
Although probiotics are considered to be safe, there are chiefly three theoretical concerns associated with the safety of probiotics, including the occurrence of disease (e.g., bacteremia or endocarditis, toxic or metabolic impacts on the gastrointestinal tract) and the transfer of antibiotic resistance in the gastrointestinal flora. Consequently, the safety of these probiotics was needed to be effectively checked before their introduction to the market for the target population’s manufacturers. Probiotics had to be safe under the intended conditions of use (4). In this regard, effective factors include the inherent properties of the microbe, the physiologic status of the consumer, the dose administered, and the possibility that probiotic bacteria could be a potential source of antibiotic resistance transferred within the gastrointestinal tract (5).
Nowadays, the interaction between probiotic bacteria and different antibiotics culminated in increasing antibiotic-resistant genes in the probiotic bacteria (5). In addition, the presence of these probiotics increases the possibility of transferring antibiotic-resistant genes to gut microflora and can be potentially transferred to pathogenic bacteria (6).
There was already an extensive set of literature regarding the antibiotic resistance of lactic acid bacteria (5). Hence, the existence of antibiotic resistance among the intestinal microbiota might be possible. Therefore, to avoid transfer-ring antibiotic-resistant genes, selecting the strains should be considered as an important act (7).It is noted that resistant gene shifts occurring at a high frequency and persisting across generations have been significantly shown in vitro and/or in vivo either from Lactobacilli to pathogens or vice versa (8).
It is noteworthy that isolates were not vulnerable to certain antibiotics such as vancomycin, tobramycin, kanamycin, and norfloxacin while they were highly vulnerable to chloramphenicol, penicillin G, erythromycin, clindamycin, and oleandomycin although none proved hemolytic activity. Accordingly, the increase of several probiotic microorganisms in dairy products and pharmaceutical formulations, and limited studies performed on the safety of probiotics should not carry any genes for antibiotic resistance transfer.
Lactobacilli spp., via OPA11 and OPD20 primers, which are used to amplify the isolated colonies by random amplified polymorphic DNA-polymerase chain reaction (PCR), has proven to be effective in discrimination, classification, and delineation of the bacterial culture and its origin clustering (9). Therefore, the present study aimed to evaluateantibiotic-resistant patterns in Lactobacillus spp. and determine the type of antibiotic resistance of chromosome- or plasmid-mediated isolates.
Methods
Isolation of Lactobacillus spp. From Dairy Products
Twenty samples, from commercial dairy products viz., cheeses, and yoghurts were collected during the first six months of 2019.To perform the experiment, 1g of the samples was added into the liquid De Man, Rogosa, and Sharpe (MRS) broth and the suspension was incubated at 37 °C in microaerophilic conditions for 24 hours. Then, a loopful of the MRS broth was streaked on the MRS agar and incubated at 37 °C for 48 hours. The isolates were identified by phenotypical tests such as catalase, oxidase, oxidative/fermentative, nitrate, and fermentation of different sugars(10).
Antibiotics Susceptibility of Lactobacillus Isolates
The disk-diffusion method was used to determine the antibiotic susceptibility of the isolates. To perform the test, the overnight growth cultures of the isolates were diluted with sterile normal saline (equal to the turbidity of a 0.5 McFarland tube). Then, the suspensions were cultivated on Mueller-Hinton agar (MHA) plates and the antibiotic disks (Patanteb, Iran) of erythromycin (15 µg), tetracycline (30 µg), vancomycin (30 µg), gentamicin (15 µg), streptomycin (10 µg), chloramphenicol (30 µg), ampicillin (10 µg), amoxicillin (25 µg), cefotaxime (30 µg), imipenem (10 µg), and ciprofloxacin (5 µg) were placed on the seeded MHA. The plates were incubated at 37°C for 48 hours and the diameters of the inhibition zones around each antibiotic disk were measured and recorded after this period (11). Antibiotic susceptibility testing results were interpreted using the Clinical Laboratory Standards Institute (CLSI) guidelines 2019.
Plasmid Isolation
The plasmids from Lactobacillus strains were isolated by the DNA plasmid extraction kit and then verified by 2% agarose gel electrophoresis in a submerged horizontal gel apparatus. The electrophoresis apparatus adjusted to 80 V for 45 minutes. Having finished the electrophoresis, the visible bands were evaluated by the alpha imager gel documentation system (Hidolgh, Germany) under ultraviolet at 260 nm(10).
Plasmid Curing
In the plasmid curing study, isolated Lactobacilli strains showing resistance to most antibiotics and a harbored plasmid band were selected for plasmid curing and put into effect. Plasmid curing was performed using acridine orange and elevated temperature (44°C) as curing agents.
To perform the experiment, 20 µL of the overnight growing culture of five antibiotic-resistant Lactobacillus strains was subjected to plasmid curing, each of which was inoculated into the tube containing 1 mL MRS broth, prepared and supplemented with different concentrations (i.e., 0.01, 0.005, 0.0025, and 0.00125 g/mL) of the acridine orange. The samples were then incubated at 37°C for 48 hours. After 48 hours of incubation, the minimal inhibitory concentration (MIC) of the acridine orange was determined, and then the sub-minimum inhibitory concentration (SIC) was considered as plasmid curing.
The elevated temperature for plasmid curing was done by the inoculation of the overnight growing culture of Lactobacillus strains into a tube containing 5 mL MRS broth and incubated at 44°C for 24 hours.
The culture with the SIC of curing agents for each Lactobacillus strain and the MRS broth incubated at 44°C were inoculated on the MRS agar and incubated at 37°C for 48 hours(10).
Antibiotic Assay of Plasmid Curing Lactobacillus Isolates
The antibiotic resistance pattern of non-cured Lactobacillus strains was selected for evaluating the antibiotic susceptibility of cured Lactobacillus isolates. Four antibiotics of vancomycin (30 µg), gentamicin (15 µg), streptomycin (10 µg), and ciprofloxacin (5 µg) were selected to perform this experiment. Simultaneously, single colonies were picked up by sterile toothpicks and inoculated on MHA plates with and without antibiotics. Each MHA plate contained only one antibiotic concentration. Then, the plates were incubated at 37°C for 48 hours. A growth colony on MHA w/wo antibiotic was considered chromosomally mediated antibiotic resistance. However, the growth colony on MHA containing no antibiotics and no growth of a given bacterium on MHA containing antibiotic was considered plasmid-mediated antibiotic resistance.
Results
Isolation of Lactobacillus spp.
The five isolated strains with desirable probiotic features were identified as Lactobacillus. These strains were L. rhamnosus, L. paracasei, L. casei, L. plantarum,and L. fermentum.
Antibiotics Susceptibility Test
The results of the antibiotic susceptibility of the isolates by the disk-diffusion method were evaluated by CLSI guidelines 2019 and expressed as sensitive (S), marginally susceptible (I), and resistant (R) as per the recommended reported standards (Table 1).
Table 1.
Antibiotic Susceptibility of Lactobacilli Isolates
|
Antibiotics
|
Susceptible (%)
|
Resistance (%)
|
Intermediate (%)
|
| Amoxicillin (25 μg) |
100 |
- |
- |
| Ampicillin (10 μg) |
37 |
20 |
7 |
| Imipenem (10 μg) |
100 |
- |
- |
| Cefotaxim (30 μg) |
60 |
33 |
7 |
| Tetracycline (30 μg) |
34 |
13 |
53 |
| Erythromycin (15 μg) |
67 |
7 |
26 |
| Chloramphenicol (30 μg) |
93 |
- |
7 |
| Vancomycin (30 μg) |
- |
100 |
- |
| Ciprofloxacin (5 μg) |
- |
100 |
- |
| Gentamicin (15 μg) |
7 |
93 |
- |
| Streptomycin (10 μg) |
13 |
87 |
- |
The results of antibiotic susceptibility indicated that all isolates were resistant to vancomycin and ciprofloxacin (100%), gentamicin (93%), and streptomycin (87%). Although all isolates were susceptible to amoxicillin and imipenem (100%), chloramphenicol (93%) was resistant to vancomycin and ciprofloxacin (100%), gentamicin (93%), and streptomycin (87%). Based on the results, all isolates were susceptible to amoxicillin and imipenem (100%), chloramphenicol (93%), and ampicillin (73%). The isolates showed different responses compared with other antibiotics.
Plasmid Isolation
As shown in Figure 1, the result of plasmid isolation indicated that all Lactobacilli strains were harbored plasmids.
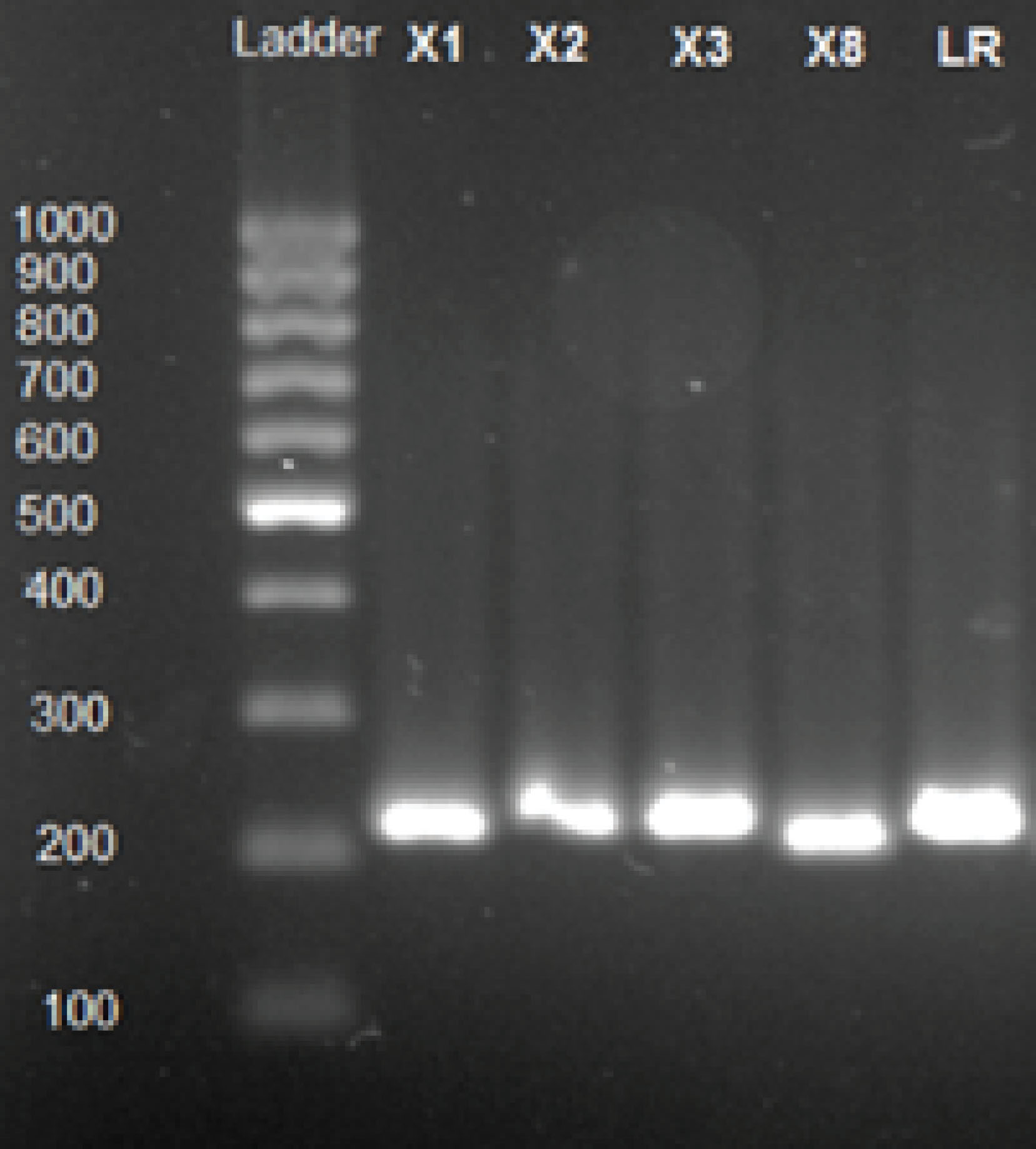
Figure 1.
Gel Electrophoresis of Plasmid DNA Isolated From Lactobacillus spp.
.
Gel Electrophoresis of Plasmid DNA Isolated From Lactobacillus spp.
Plasmid Curing
Plasmid curing experiments on tested Lactobacilli strains were performed using both the curing agent of the acridine orange and elevated temperature (44°C). The results of the MIC and SIC of acridine orange for Lactobacilli strains (Table 2) revealed that Lactobacillus plantarum was relatively more resistant to acridine orange.
Table 2.
MIC and SIC of Acridine Orange Concentration Against Antibiotic-resistant Lactobacillus Isolates
|
Strains
|
MIC
mg/mL
|
SIC
mg/mL |
|
Lactobacillus rhamnosus
|
0.00125 |
0.0025 |
|
Lactobacillus paracasei
|
0.00125 |
0.0025 |
|
Lactobacillus casei
|
0.00125 |
0.0025 |
|
Lactobacillus fermentum
|
0.0025 |
0.005 |
|
Lactobacillus plantarum
|
0.0025 |
0.005 |
Note. MIC: Minimal inhibitory concentration; SIC: Subminimal inhibitory concentration.
Antibiotic Assay of Plasmid Curing Lactobacillus Isolates
The obtained results from antibiotic susceptibility after plasmid curing represented that all L. isolates, L. rhamnosus, and Lactobacillus plantarum lost resistance phenotypically to streptomycin and gentamicin. Hence, streptomycin and gentamicin-resistant properties were of plasmid-mediated type in these two strains. However, vancomycin and ciprofloxacin-resistant properties were of chromosomally-mediated type.
Discussion
Although several microorganisms such as Lactobacillus, Bifidobacteria, Enterococcus, and Streptococcus are introduced as probiotic bacteria, (12,13),information on their antibiotic-resistant characters is not widely available yet (14). In this study, five Lactobacillus strains were isolated from commercial products and assessed for their susceptibility to different antibiotics. The isolated strains were resistant to vancomycin and ciprofloxacin (100%) in addition to gentamicin (93%) and streptomycin (87%) while they were susceptible to amoxicillin and imipenem (100%), as well as chloramphenicol (93%) and ampicillin (73%). However, all the isolates harbored DNA plasmid although the plasmid-mediatedresistant character was found only in two strains. This means that streptomycin and gentamicin resistance genes in L. rhamnosus and L. plantarum are located on the plasmid, and therefore, horizontal gene transfer among probiotic strains might be expectable. Comunian et al (15) isolated 197 strains of lactobacilli from Italy’s fermented products. They verified the results by observing their resistance genes through the PCR method (13)and found that all strains were resistant to erythromycin and tetracycline. Zheng et al reported that 88% of lactic acid bacteria isolates were resistant to chloramphenicol and tetracycline. In addition, they found that the isolated strains could transfer antibiotic-resistance gene tet(M) to pathogenic bacteria in the human gut (16).
Furthermore, several reports demonstrated that many probiotics such as lactobacilli were resistant to vancomycin, gentamicin, streptomycin, and ciprofloxacin, and most were of plasmid-mediated type (16,17).
On the other hand, the findings of several studies in Italy and Spain showed that some probiotic bacteria such as L. rhamnosus and L. jensenii have potent activity for causing bacteremia (15). Therefore, the possibility of transferring antibiotic-resistant genes and pathogenic potential of some probiotics should be considered as a critical option for selecting probiotic microorganisms. Hence, as mentioned above, the antibiotic resistance character in probiotic bacteria has some advantages and disadvantages.
Survival in people on oral antibiotic therapy is an advantage while acquired resistance genes within the bacterial species of gut microflora or to pathogenic commensals is a disadvantage. Therefore, to introduce commercial probiotic bacteria, several standard screenings including their evaluation for carriage antibiotic resistance genes should be done to eliminate the distribution of the genes among various gut microflora and potentially pathogenic bacteria.
Conflict of Interests
None.
Acknowledgements
The authors thank all who helped us at the Department of Microbiology, Kazeroon Branch, Islamic Azad University, Iran.
References
- Amara AA, Shibl A. Role of Probiotics in health improvement, infection control and disease treatment and management. Saudi Pharm J 2015; 23(2):107-14. doi: 10.1016/j.jsps.2013.07.001 [Crossref] [ Google Scholar]
- del Campo R, Garriga M, Pérez-Aragón A, Guallarte P, Lamas A, Máiz L. Improvement of digestive health and reduction in proteobacterial populations in the gut microbiota of cystic fibrosis patients using a Lactobacillus reuteri probiotic preparation: a double blind prospective study. J Cyst Fibros 2014; 13(6):716-22. doi: 10.1016/j.jcf.2014.02.007 [Crossref] [ Google Scholar]
- Matussin NB, Chin YY, Ishan I, Shivanand P. Fermented food in Asia as a source of potential probiotics: properties and beneficial effects. Scientia Bruneiana 2016; 15:18-32. doi: 10.46537/scibru.v15i0.40 [Crossref] [ Google Scholar]
- Hirayama K, Rafter J. The role of probiotic bacteria in cancer prevention. Microbes Infect 2000; 2(6):681-6. doi: 10.1016/s1286-4579(00)00357-9 [Crossref] [ Google Scholar]
- Singh K, Kallali B, Kumar A, Thaker V. Probiotics: a review. Asian Pac J Trop Biomed 2011; 1(2):S287-90. doi: 10.1016/s2221-1691(11)60174-3 [Crossref] [ Google Scholar]
- Sharma P, Tomar SK, Goswami P, Sangwan V, Singh R. Antibiotic resistance among commercially available probiotics. Food Res Int 2014; 57:176-95. doi: 10.1016/j.foodres.2014.01.025 [Crossref] [ Google Scholar]
- Beige F, Baseri Salehi M, Bahador N, Mobasherzadeh S. Plasmid mediated antibiotic resistance in isolated bacteria from burned patients. Jundishapur J Microbiol 2015; 8(1):e13567. doi: 10.5812/jjm.13567 [Crossref] [ Google Scholar]
- Stanton C, Gardiner G, Meehan H, Collins K, Fitzgerald G, Lynch PB. Market potential for probiotics. Am J Clin Nutr 2001; 73(2 Suppl):476S-83S. doi: 10.1093/ajcn/73.2.476s [Crossref] [ Google Scholar]
- Homayouni A, Alizadeh M, Alikhah H, Zijah V. Functional Dairy Probiotic Food Development: Trends, Concepts, and Products. Rijeka, Croatia: IntechOpen; 2012.
- Baserisalehi M, Bahador N. A study on relationship of plasmid with antibiotic resistance in thermophilic Campylobacter spp isolates from environmental samples. Biotechnology 2008; 7(4):813-7. doi: 10.3923/biotech.2008.813.817 [Crossref] [ Google Scholar]
- Pyar H, Peh KK. Characterization and identification of Lactobacillus acidophilus using biolog rapid identification system. Int J Pharm Pharm Sci 2014; 6(1):189-93. [ Google Scholar]
- Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 2004; 12(9):412-6. doi: 10.1016/j.tim.2004.07.004 [Crossref] [ Google Scholar]
- Fouhy F, Stanton C, Cotter PD, Hill C, Walsh F. Proteomics as the final step in the functional metagenomics study of antimicrobial resistance. Front Microbiol 2015; 6:172. doi: 10.3389/fmicb.2015.00172 [Crossref] [ Google Scholar]
- del Carmen Casado Muñoz M, Benomar N, Ennahar S, Horvatovich P, Lavilla Lerma L, Knapp CW. Comparative proteomic analysis of a potentially probiotic Lactobacillus pentosus MP-10 for the identification of key proteins involved in antibiotic resistance and biocide tolerance. Int J Food Microbiol 2016; 222:8-15. doi: 10.1016/j.ijfoodmicro.2016.01.012 [Crossref] [ Google Scholar]
- Comunian R, Daga E, Dupré I, Paba A, Devirgiliis C, Piccioni V. Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int J Food Microbiol 2010; 138(1-2):151-6. doi: 10.1016/j.ijfoodmicro.2009.11.018 [Crossref] [ Google Scholar]
- Zheng M, Zhang R, Tian X, Zhou X, Pan X, Wong A. Assessing the risk of probiotic dietary supplements in the context of antibiotic resistance. Front Microbiol 2017; 8:908. doi: 10.3389/fmicb.2017.0090 [Crossref] [ Google Scholar]
- Dinu LD, Babata SI, Ciurea AN, Lunita AM. Antimicrobial effect and antibiotic resistance of lactic acid bacteria from some commercial dairy products. Scientific Papers: Animal Science & Biotechnologies 2018; 51(1):114-8. [ Google Scholar]