Avicenna Journal of Clinical Microbiology and Infection. 11(3):113-118.
doi: 10.34172/ajcmi.3562
Original Article
Investigating the Presence of blaCMY and Extended Spectrum Beta-Lactamase Genes in Klebsiella pneumoniae Isolates Identified in the Clinical Samples of Patients in Dhi-Qar, Iraq
Abeer Taleb Ali 1  , Seyedeh Elham Rezatofighi 1, *
, Seyedeh Elham Rezatofighi 1, *  , Hossein Motamedi 1, *
, Hossein Motamedi 1, *  , Mohammad Roayaei Ardakani 1
, Mohammad Roayaei Ardakani 1
Author information:
1Department of Biology, Faculty of Science, Shahid Chamran University of Ahvaz, Ahvaz, Iran
Abstract
Background: Klebsiella pneumoniae is an opportunistic pathogen that can cause nosocomial infections due to its high virulence factors and multiple antimicrobial resistance (AMR) mechanisms. This bacterium can produce various beta-lactamases, including extended-spectrum beta-lactamase (ESBL) and AmpC. This study aimed to investigate the presence of plasmid AmpC and ESBL genes in K. pneumoniae isolates from the clinical samples of patients in Dhi-Qar, Iraq.
Methods: A total of 612 clinical samples were collected from different medical centers and laboratories in Dhi-Qar, Iraq, between April 2023 and February 2024. Then, the presence of K. pneumoniae in these samples was evaluated using conventional biochemical and microbiological methods. ESBL production was assessed phenotypically by the synergy double disk (SDD) method. The presence of the blaCTX-M, blaTEM, blaCTX-M-3, blaCMY, and blaSHV genes was analyzed using the polymerase chain reaction.
Results: Out of the 612 samples, 180 (29.4%) tested positive for K. pneumoniae. Of these, 40 isolates (22.2%) were positive in the SDD test and were considered ESBL producers. The blaSHV, blaCTX-M-3, blaTEM, and blaCTX-M genes were detected in 8 (20%), 32 (80%), 21 (52.5%), and 22 (55%) isolates, respectively. The blaCMY gene was not found in any of the K. pneumoniae isolates.
Conclusion: Our study highlights high resistance against third-generation cephalosporins among K. pneumoniae isolates. The prevalence of ESBL genes, including blaCTX-M, blaTEM, and blaSHV, among these isolates can cause serious challenges in the treatment of bacterial infections.
Keywords: Klebsiella pneumoniae, AmpC, Beta-Lactamase, Synergy double disk, Extended-spectrum beta-lactamase
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Taleb Ali A, Rezatofighi SE, Motamedi H, Roayaei Ardakani M. Investigating the presence of blaCMY and extended spectrum beta-lactamase genes in Klebsiella pneumoniae isolates identified in the clinical samples of patients in Dhi-Qar, Iraq. Avicenna J Clin Microbiol Infect. 2024; 11(3):113-118. doi:10.34172/ajcmi.3562
Introduction
Klebsiella pneumonia, a member of the Enterobacteriaceae family, is an opportunistic bacterium capable of causing different diseases such as urinary tract infections (UTIs), pneumoniae, septicemia, and soft tissue infections, particularly in hospitals (1). It accounts for 3%–8% of all nosocomial bacterial infections (2). K. pneumoniae expresses several virulence factors, including endotoxins, siderophores, adhesins, iron-scavenging mechanisms, and capsules (3).This is a serious threat to human health, making antimicrobial resistance (AMR), a major global infection resistant to third-generation cephalosporins (4). Health systems currently incur significant financial expenditures due to AMR (5). The prevalence and diversity of AMR genes make them one of the major challenges facing global healthcare systems (5).
Beta-lactam antibiotics are commonly consumed to treat diseases caused by Enterobacteriaceae. However, the loss of sensitivity to these antimicrobial agents in Gram-negative bacteria, particularly Enterobacteriaceae, is spreading rapidly worldwide. Resistance to beta-lactams is primarily attributed to the production of three types of beta-lactamases (BLs), namely, extended-spectrum beta-lactamase (ESBL), AmpC-type BLs (AmpC-BLs), and carbapenemases (6).
ESBLs are a complex, diverse, and quickly evolving category of enzymes that pose significant challenges in treating patients with both community- and hospital-acquired infections (7). Most ESBLs are plasmid-mediated. Members of the Enterobacteriaceae family can easily transfer these plasmids among themselves, leading to the accumulation of resistance genes and the production of strains with multidrug-resistant plasmids. Isolates that produce ESBLs are resistant to several antibiotic classes. Unfortunately, plasmids that generate ESBLs are relatively stable in the host bacterium (8).
Based on Ambler classification, ESBL enzymes are categorized into two A and D classes. ESBLs, or serine beta-lactamases, form the largest group of beta-lactamases (9). The most prevalent genes encoding enzymes in class A include blaTEM, blaCTX-M, and blaSHV (10). Before the 2000s, TEM and SHV were the most widespread ESBLs, but later, CTX-M became the most frequent worldwide, leading to a gradual decline in TEM-type ESBLs. Most CTX-M enzymes are more effective on cefotaxime than ceftazidime (9).
AmpC BLs are able to hydrolyze broad-spectrum cephalosporins as well as penicillins. AmpC-BLs belong to Ambler Class C and can be encoded either by the chromosome or plasmids. Plasmid-encoded AmpCs (pAmpCs) are widely spread and encoded by different genes, among which blaCMY is the most prevalent. The pAmpC genes can be disseminated through K. pneumoniae and Escherichia coli, the bacteria responsible for diverse hospital-acquired infections (11).
Our research attempted to evaluate the presence of two types of BLs, ESBL and pAmpC, in K. pneumoniae isolates from the clinical samples of patients in Dhi-Qar, Iraq.
Materials and Methods
Sample Collection and Identification
In total, 612 samples were gathered from individuals suffering from blood septicemia, respiratory tract infections, burns, wound infections, and UTIs. The samples were gathered from various hospitals in Dhi-Qar, Iraq, including Al-Hussein hospital, Al Nasiriya hospital, Al Haboubi hospital, Al-Musawi Children’s hospital, Bint Al Huda hospital, and medical laboratories in the city of Nasiriyah from April 2023 to February 2024. The samples were cultivated on MacConkey agar, blood agar, and nutrient agar. All culture media were obtained from Merck Company (Germany). Morphological characteristics of colonies, such as shape, size, margins, and pigmentation, were investigated in this study. Grown colonies were identified using Gram staining and traditional biochemical and microbiological tests, including oxidase, catalase, IMViC, sulfur indole motility, and triple sugar iron agar.
Synergy Double Disk Method for the Detection of ESBL Isolates
To identify ESBL-producing isolates, ceftazidime- or cefotaxime-resistant isolates were evaluated using the SDD test, as recommended by CLSI 2023 (12). The ceftazidime (CAZ; 30 μg), cefotaxime (CTX; 30 μg), ceftazidime-clavulanic acid (TZL; 30-10 μg), and cefotaxime-clavulanic acid (CEC; 30-10 μg) disks were applied in this method. The test isolates were inoculated onto Muller-Hinton agar (Merck, Germany) plates, and the disks were placed 3.5 cm from each other. The diameter of the no-growth zones around the disks was measured after 16 hours of incubation. According to CLSI criteria, the isolates were recorded as ESBL producers if the diameter of the no-growth zone around ceftazidime-clavulanic acid or cefotaxime-clavulanic acid (combined disks) increased by ≥ 5 mm compared to ceftazidime or cefotaxime (individual disks).
DNA Extraction
A manual boiling process was applied to extract DNA from K. pneumoniae isolates (13). A single colony from MacConkey agar was picked with an inoculation loop and cultured into the nutrient broth, then incubated for 18 hours at 37 °C. After incubation, 200 μL of the cultured media was transferred to a 1.5 mL microtube and centrifuged for 5 minutes at 5000 rpm. Afterward, the supernatant was discarded, and 200 μL of water was added to the bacterial pellet. After pipetting, the suspensions were boiled for 10 minutes and then centrifuged for 5 minutes at 10 000 rpm. The supernatant was collected as the template for amplification and stored at -20 °C.
Detection of Extended-Spectrum Beta-Lactamase Genes and blaCMY
The test isolates were evaluated for the presence of four ESBL genes, including blaTEM, blaCTX-M-3, blaSHV, and blaCTX-M, along with the blaCMY gene related to the pAmpC-BL type. The primer sequences are provided in Table 1. The polymerase chain reaction conditions were applied as previously described (14-18).
Table 1.
The Primer Sequences for the Detection of ESBL Genes andblaCMY
|
Gene
|
Sequences
|
Annealing Temperature (°C)
|
Product Size
|
Reference
|
|
bla
TEM
|
F: ATAAAATTCTTGAAGACGAAA
R: GACAGTTACCAATGCTTAATC |
56 |
1080 |
(14) |
|
bla
CTX-M
|
F: CGATGTGCAGTACCAGTAA
R: TTAGTGACCAGAATCAGCGG |
60 |
585 |
(15) |
|
bla
CTX-M-3
|
F: CGCTTTGCGATGTGCAG
R: ACCGCGATATCGTTGGT |
60 |
550 |
(16) |
|
bla
CMY
|
F: ATGATGAAAAAATCGTTATGCT
R: TTATTGCAGCTTTTCAAGAATGCG |
60 |
1140 |
(17) |
|
bla
SHV
|
F: CACTCAAGGATGTATTGTG
R: TTAGCGTTGCCAGTGCTCG |
50 |
885 |
(18) |
Note. ESBL: Extended-spectrum beta-lactamase.
Statistical Analysis
The χ2 or Fisher’s exact test was applied to analyze descriptive data, and a P value of ≤ 0.05 was considered statistically significant.
Results
Out of 612 different samples, 180 (29.4%) were positive for K. pneumonia, including urine, burn, sputum, wound, blood, and pulmonary fluid samples. Details are included in Table 2. Of the positive samples, 48.3% (n = 87) and 51.7% (n = 93) were from females and males, respectively (P> 0.05).
Table 2.
The Number of Klebsiella pneumonia Isolates From Different Clinical Samples
|
Sample
|
Urine
|
Burn
|
Wound
|
Sputum
|
Blood
|
Pulmonary Fluids
|
| No. (%) |
59 (32.8) |
8 (4.4) |
22 (12.2) |
68 (37.8) |
13 (7.2) |
10 (5.6) |
|
Genus
|
M
|
F
|
M
|
F
|
M
|
F
|
M
|
F
|
M
|
F
|
M
|
F
|
| N (%) |
26 (44.1) |
33 (55.9) |
5 (62.5) |
3 (37.5) |
10 (45.4) |
12 (54.6) |
40 (58.8) |
28 (41.2) |
9 (69.2) |
4 (30.8) |
3 (30) |
7 (70) |
Note. M: Male; F: Female; N: Number.
Phenotypic Detection of Extended-Spectrum Beta-Lactamase- Producing Isolates
AMR was observed against cefotaxime, ceftazidime, or both in 139 (77.2%), 117 (65%), and 95 (58%) of K. pneumoniae isolates, respectively. After subjecting these isolates to the SDD test, 40 (22.2%) were detected as ESBL producers.
Detection of Extended-Spectrum Beta-Lactamase Genes and blaCMY
In total, eight (20%), 32 (80%), 22 (55%), and 21 (52.5%) isolates harbored blaSHV, blaCTX-M-3,blaCTX-M, andblaTEM. The blaCMY was found in none of the K. pneumoniae isolates (Figure 1). Four (10%) isolates did not exhibit any of the tested genes. Some isolates harbored multiple ESBL genes, with eight (20%) containing two ESBL genes and five (12.5%) consisting of three ESBL genes. The frequency of ESBL gene profiles is provided in Table 3.
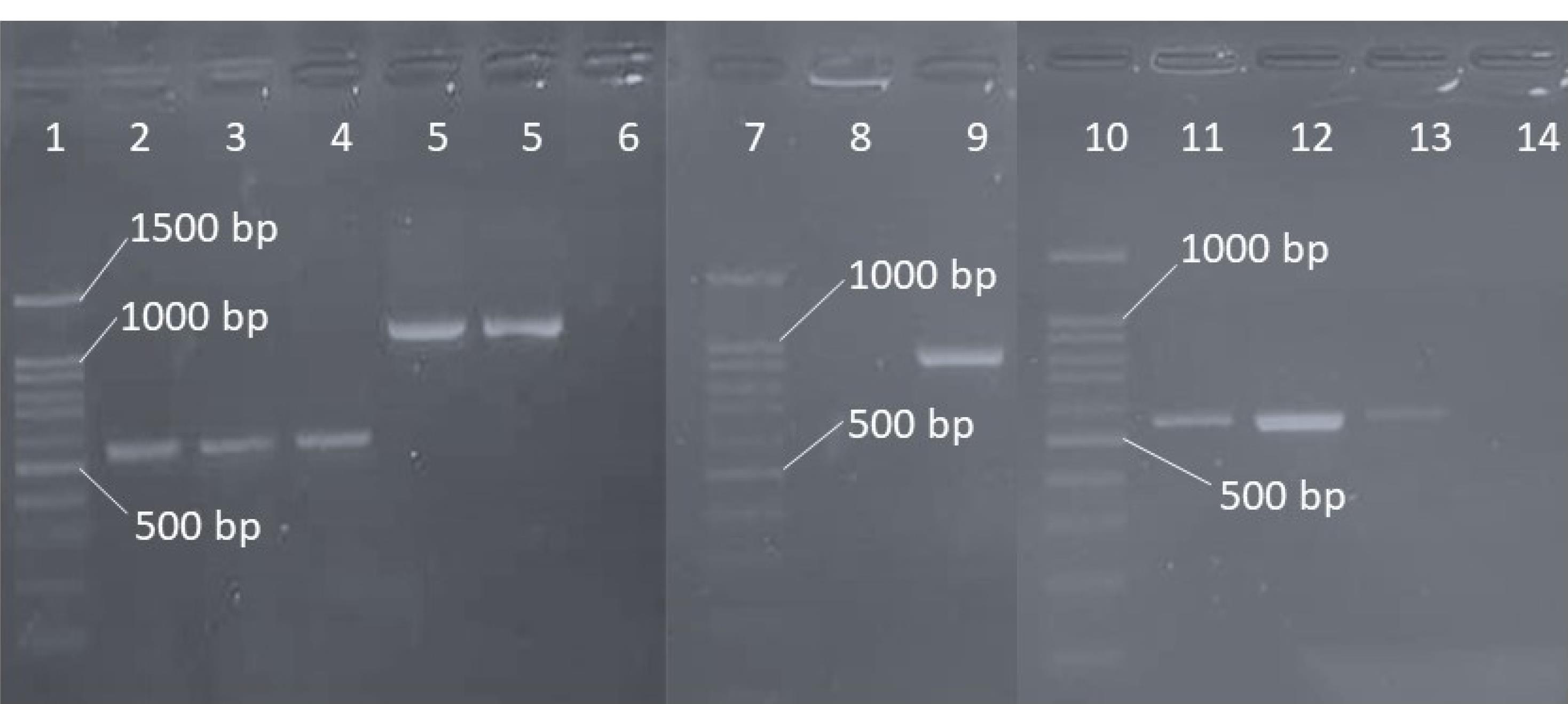
Figure 1.
PCR Results of ESBL Genes. Note. PCR: Polymerase chain reaction; ESBL: Extended-spectrum beta-lactamase. Wells 1, 8, and 11: Ladder (100 bp); Wells 2, 3, and 4: blaCTX-M-3; Wells 5 and 6: blaTEM; Well 10: blaSHV; Wells 12, 13, and 14: blaCTX-M; Wells 7, 9, and 15: Negative isolates
.
PCR Results of ESBL Genes. Note. PCR: Polymerase chain reaction; ESBL: Extended-spectrum beta-lactamase. Wells 1, 8, and 11: Ladder (100 bp); Wells 2, 3, and 4: blaCTX-M-3; Wells 5 and 6: blaTEM; Well 10: blaSHV; Wells 12, 13, and 14: blaCTX-M; Wells 7, 9, and 15: Negative isolates
Table 3.
The ESBL Gene Profiles Found Among Klebsiella pneumoniae Isolates
|
Genes
|
bla
SHV
N (%)
|
bla
CTX-M-3
N (%)
|
bla
CTX-M
N (%)
|
bla
TEM
N (%)
|
bla
CMY
N (%)
|
bla
CTX-M-3
-
bla
TEM
N (%)
|
bla
CTX-M-3
-
bla
SHV
N (%)
|
bla
CTX-M-3
or
bla
CTX-M
-
bla
TEM
N (%)
|
bla
CTX-M-3
-
bla
SHV
-
bla
TEM
N (%)
|
No Gene
N (%)
|
| Sum |
8 (20) |
32 (80) |
22 (55) |
21 (52.5) |
0 (0) |
4 (10) |
1 (2.5) |
7 (17.5) |
5 (12.5) |
4 (10) |
Discussion
Enterobacteriaceae, especially K. pneumonia,cause nosocomial infections. Recently, AMR, due to the presence of BL, has increased worldwide, leading to resistance against a wide variety of antimicrobial agents (19). The prevalence of beta-lactamase producers and related genes varies greatly across different regions and from one year to another. For the accurate detection of ESBL-producing microorganisms, genotypic tests are required in addition to phenotypic tests (20).
The results of our study revealed that blaCTX-M-3 and blaCTX-M were the most frequent ESBL genes among K. pneumoniae isolates, respectively. Multiple studies have reported that CTX-M–type ESBLs are probably the most common ESBL type worldwide (7). CTX-M has different types, among which CTX-M-3 BL is the most common ESBL expressed by K. pneumoniae, E. coli, and different serotypes of non-typhoid Salmonella (21-23). The universal primers of blaCTX-M and specific primers were used for blaCTX-M-3 because, in some cases, universal primers cannot detect all blaCTX-M-3 genes. Only one isolate was negative for blaCTX-M-3 but positive for blaCTX-M, demonstrating that approximately 95% of blaCTX-M-positive isolates harbored blaCTX-M-3. These isolates may carry other types of blaCTX-M simultaneously.
The frequency rate of blaCTX-M varies in different geographic areas of Iraq. Studies conducted in Baghdad (24), Najaf (25), and Erbil (26) reported frequencies of 90%, 88.2%, and 41.1% for blaCTX-M, respectively. Raouf et al found a prevalence of 47.4% for blaCTX-M among K. pneumoniae isolates of patients with community-acquired pneumonia (27). In our study, blaTEM was observed in 52.5% of ESBL-producing isolates. In some studies performed in Iraq, this gene was the most prevalent ESBL gene (24,26). The frequency of blaTEM in Baghdad (24), Erbil (26), and Sulaimani (20) was 95%, 64.7%, and 53.7%, respectively. blaSHV, another important ESBL gene, was observed in 20% of ESBL-producing isolates. Different frequencies ranging from 15.8% (27) to 92.85% (20) have been reported in other studies. The results of our investigation differed from those of some other studies, suggesting that different geographical regions may have various prevalence rates and types of ESBL genes. In addition, the sample size, type of sample, year of sampling, and method used for gene detection may have affected the study results.
The findings related to the prevalence of ESBL genes outside of Iraq also vary widely. In a study conducted by Saisi et al, the most common genes responsible for producing ESBLs were CTX-M (100%), TEM (97%), and SHV (94%) in Kenya, respectively (28). CTX-M was found to be the predominant gene according to studies from South America, the UK, Spain, the USA, and numerous regions of the Indian subcontinent (29). The blaTEM gene was confirmed to be more prevalent than SHV in a Chinese investigation (30).
The presence of more than one BL gene within each isolate occurred in 13 ESBL-producing isolates. Notably, most isolates had a double or triple combination of ESBL genes, which is probably due to their transport by a common plasmid. Such plasmids can usually carry other resistance genes, leading to resistance against other antimicrobial categories. In Tunisia, Alibi et al found a triple combination of SHV/TEM/CTX-M (31). In Tanzania, Mshana et al reported a combination of CTX-M/TEM in 11.96% and SHV/CTX-M in 10.87% of the isolates (32).
Based on the results, the blaCMY gene was not observed among cefotaxime- or ceftazidime-resistant isolates. There have been limited studies on the worldwide distribution of pAmpC-BLs, including blaCMY, compared to the more frequently reported carbapenemase- and ESBL-producing bacteria (33-35). In general, the lowest frequency was found in Europe, including 0.06% in Denmark (36), 2.6% in Holland (37), and 11.9% in Germany (38), followed by America, with rates ranging from 1.3% in 2016 (37) to 3.42% in 2019 (39).
However, the prevalence of this type of BL is higher in the Middle East and Asia than in the rest of the world, especially in Iran (20.50% in 2020) (40), China (31.5% in 2015) (41), and Nepal (40.26% in 2020) (42). It was impossible to find information on the frequency of blaCMY among K. pneumoniae strains isolated from patients in Iraq.
Among identified ESBL-producing K. pneumoniae isolates, 10% were negative for the investigated genes. ESBL production in these isolates may be attributed to other ESBL genes, such as blaOXA, or other beta-lactamase groups, including metallo-beta-lactamase, carbapenemase, or other serine beta-lactamases.
Conclusion
Klebsiella pneumoniae isolates detected in our study showed a high level of resistance against third-generation cephalosporins. ESBL genes, including blaTEM, blaCTX-M, and blaSHV, were frequent among these isolates, which can lead to serious challenges in the treatment of bacterial infections. The threat posed by this group of AMR bacteria to public health should not be underestimated. Extensive studies and the development of alternative solutions are needed to address this issue.
Acknowledgments
This study has been extracted from a PhD thesis by Abeer Taleb Ali submitted to and approved by the Shahid Chamran University of Ahvaz. The authors are highly thankful to the Shahid Chamran University of Ahvaz for the facilities to accomplish the present research project within time.
Authors’ Contribution
Conceptualization: Seyedeh Elham Rezatofighi, Hossein Motamedi, Abeer Taleb Ali, and Mohammad Roayaei Ardakani.
Data curation: Seyedeh Elham Rezatofighi and Hossein Motamedi.
Formal analysis: Seyedeh Elham Rezatofighi, Hossein Motamedi, Abeer Taleb Ali, and Mohammad Roayaei Ardakani.
Investigation: Abeer Taleb Ali.
Methodology: Abeer Taleb Ali, Seyedeh Elham Rezatofighi, Hossein Motamedi, and Mohammad Roayaei Ardakani.
Project administration: Seyedeh Elham Rezatofighi and Hossein Motamedi.
Resources: Abeer Taleb Ali, Seyedeh Elham Rezatofighi, Hossein Motamedi, and Mohammad Roayaei Ardakani.
Supervision: Seyedeh Elham Rezatofighi and Hossein Motamedi.
Validation: Seyedeh Elham Rezatofighi and Hossein Motamedi.
Visualization: Seyedeh Elham Rezatofighi and Hossein Motamedi.
Writing–original draft: Abeer Taleb Ali, Seyedeh Elham Rezatofighi, Hossein Motamedi, and Mohammad Roayaei Ardakani.
Writing–review & editing: Seyedeh Elham Rezatofighi and Hossein Motamedi.
Competing Interests
The author declared no conflict of interests.
Ethical Approval
The ethical permission of the present study was taken from the Ethics Committee of Shahid Chamran University of Ahvaz according to the Declaration of Helsinki (IR.SCU.REC.1403.073).
Funding
There is no funding or support for the publication of this study.
References
- Jondle CN, Gupta K, Mishra BB, Sharma J. Klebsiella pneumoniae infection of murine neutrophils impairs their efferocytic clearance by modulating cell death machinery. PLoS Pathog 2018; 14(10):e1007338. doi: 10.1371/journal.ppat.1007338 [Crossref] [ Google Scholar]
- Aghamohammad S, Badmasti F, Solgi H, Aminzadeh Z, Khodabandelo Z, Shahcheraghi F. First report of extended-spectrum betalactamase-producing Klebsiella pneumoniae among fecal carriage in Iran: high diversity of clonal relatedness and virulence factor profiles. Microb Drug Resist 2020; 26(3):261-9. doi: 10.1089/mdr.2018.0181 [Crossref] [ Google Scholar]
- Rønning TG, Aas CG, Støen R, Bergh K, Afset JE, Holte MS. Investigation of an outbreak caused by antibiotic-susceptible Klebsiella oxytoca in a neonatal intensive care unit in Norway. Acta Paediatr 2019; 108(1):76-82. doi: 10.1111/apa.14584 [Crossref] [ Google Scholar]
- Esposito EP, Cervoni M, Bernardo M, Crivaro V, Cuccurullo S, Imperi F. Molecular epidemiology and virulence profiles of colistin-resistant Klebsiella pneumoniae blood isolates from the hospital agency “Ospedale dei Colli,” Naples, Italy. Front Microbiol 2018; 9:1463. doi: 10.3389/fmicb.2018.01463 [Crossref] [ Google Scholar]
- Walter J, Haller S, Quinten C, Kärki T, Zacher B, Eckmanns T. Healthcare-associated pneumonia in acute care hospitals in European Union/European Economic Area countries: an analysis of data from a point prevalence survey, 2011 to 2012. Euro Surveill 2018; 23(32):1700843. doi: 10.2807/1560-7917.Es.2018.23.32.1700843 [Crossref] [ Google Scholar]
- Rodríguez-Guerrero E, Callejas-Rodelas JC, Navarro-Marí JM, Gutiérrez-Fernández J. Systematic review of plasmid AmpC type resistances in Escherichia coli and Klebsiella pneumoniae and preliminary proposal of a simplified screening method for AmpC. Microorganisms 2022; 10(3):611. doi: 10.3390/microorganisms10030611 [Crossref] [ Google Scholar]
- Rawat D, Nair D. Extended-spectrum β-lactamases in gram-negative bacteria. J Glob Infect Dis 2010; 2(3):263-74. doi: 10.4103/0974-777x.68531 [Crossref] [ Google Scholar]
- Rolbiecki D, Harnisz M, Korzeniewska E, Buta M, Hubeny J, Zieliński W. Detection of carbapenemase-producing, hypervirulent Klebsiella spp in wastewater and their potential transmission to river water and WWTP employees. Int J Hyg Environ Health 2021; 237:113831. doi: 10.1016/j.ijheh.2021.113831 [Crossref] [ Google Scholar]
-
Akhtar A, Fatima N, Khan HM. β-lactamases and their classification: an overview. In: Shahid M, Singh A, Sami H, eds. Beta-Lactam Resistance in Gram-Negative Bacteria: Threats and Challenges. Singapore: Springer; 2022. p. 25-33. doi: 10.1007/978-981-16-9097-6_3.
- Teban-Man A, Farkas A, Baricz A, Hegedus A, Szekeres E, Pârvu M. Wastewaters, with or without hospital contribution, harbour MDR, carbapenemase-producing, but not hypervirulent Klebsiella pneumoniae. Antibiotics (Basel) 2021; 10(4):361. doi: 10.3390/antibiotics10040361 [Crossref] [ Google Scholar]
- Rao A, Naha S, Bhattacharjee A, Chattopadhyay P, Dutta S, Basu S. Plasmid-mediated AmpC in Klebsiella pneumoniae and Escherichia coli from septicaemic neonates: diversity, transmission and phenotypic detection. J Glob Antimicrob Resist 2023; 34:9-14. doi: 10.1016/j.jgar.2023.05.012 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 33rd ed. CLSI Supplement M100. CLSI; 2023.
- Yousefipour M, Rezatofighi SE, Roayaei Ardakani M. Detection and characterization of hybrid uropathogenic Escherichia coli strains among E coli isolates causing community-acquired urinary tract infection. J Med Microbiol 2023; 72(2):001660. doi: 10.1099/jmm.0.001660 [Crossref] [ Google Scholar]
- Mabilat C, Goussard S, Sougakoff W, Spencer RC, Courvalin P. Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum beta-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 1990; 23(1):27-34. doi: 10.1016/0147-619x(90)90041-a [Crossref] [ Google Scholar]
- Batchelor M, Hopkins K, Threlfall EJ, Clifton-Hadley FA, Stallwood AD, Davies RH. blaCTX-M genes in clinical Salmonella isolates recovered from humans in England and Wales from 1992 to 2003. Antimicrob Agents Chemother 2005; 49(4):1319-22. doi: 10.1128/aac.49.4.1319-1322.2005 [Crossref] [ Google Scholar]
- Poirel L, Naas T, Le Thomas I, Karim A, Bingen E, Nordmann P. CTX-M-type extended-spectrum beta-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob Agents Chemother 2001; 45(12):3355-61. doi: 10.1128/aac.45.12.3355-3361.2001 [Crossref] [ Google Scholar]
- Kojima A, Ishii Y, Ishihara K, Esaki H, Asai T, Oda C. Extended-spectrum-beta-lactamase-producing Escherichia coli strains isolated from farm animals from 1999 to 2002: report from the Japanese Veterinary Antimicrobial Resistance Monitoring Program. Antimicrob Agents Chemother 2005; 49(8):3533-7. doi: 10.1128/aac.49.8.3533-3537.2005 [Crossref] [ Google Scholar]
- Pitout JD, Thomson KS, Hanson ND, Ehrhardt AF, Moland ES, Sanders CC. β-lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob Agents Chemother 1998; 42(6):1350-4. doi: 10.1128/aac.42.6.1350 [Crossref] [ Google Scholar]
- Besharati Zadeh S, Shakib P, Zolfaghari MR, Farajzadeh Sheikh A. Prevalence of Escherichia coli and Klebsiella pneumoniae, producing extended-spectrum beta-lactamase (ESBLs) from clinical specimen in Khuzestan, Iran. Gene Cell Tissue 2021; 8(3):e112377. doi: 10.5812/gct.112377 [Crossref] [ Google Scholar]
- Mohammed AB, Anwar KA. Phenotypic and genotypic detection of extended spectrum beta lactamase enzyme in Klebsiella pneumoniae. PLoS One 2022; 17(9):e0267221. doi: 10.1371/journal.pone.0267221 [Crossref] [ Google Scholar]
- Liu SY, Su LH, Yeh YL, Chu C, Lai JC, Chiu CH. Characterisation of plasmids encoding CTX-M-3 extended-spectrum beta-lactamase from Enterobacteriaceae isolated at a university hospital in Taiwan. Int J Antimicrob Agents 2007; 29(4):440-5. doi: 10.1016/j.ijantimicag.2006.11.013 [Crossref] [ Google Scholar]
- Su LH, Chiu CH, Chu C, Ou JT. Antimicrobial resistance in nontyphoid Salmonella serotypes: a global challenge. Clin Infect Dis 2004; 39(4):546-51. doi: 10.1086/422726 [Crossref] [ Google Scholar]
- Pai H, Choi EH, Lee HJ, Hong JY, Jacoby GA. Identification of CTX-M-14 extended-spectrum beta-lactamase in clinical isolates of Shigella sonnei, Escherichia coli, and Klebsiella pneumoniae in Korea. J Clin Microbiol 2001; 39(10):3747-9. doi: 10.1128/jcm.39.10.3747-3749.2001 [Crossref] [ Google Scholar]
- Samanje J, Mohammed AS, Al-Hamami MS. Phenotypic and genotypic detection of extended-spectrum β-lactamase production by Klebsiella pneumoniae isolated from different clinical samples in Baghdad, Iraq. J Pure Appl Microbiol 2021; 15(3):1681-8. doi: 10.22207/jpam.15.3.64 [Crossref] [ Google Scholar]
- Majeed HT, Aljanaby AA. Antibiotic susceptibility patterns and prevalence of some extended spectrum β-lactamases genes in gram-negative bacteria isolated from patients infected with urinary tract infections in Al-Najaf city, Iraq. Avicenna J Med Biotechnol 2019; 11(2):192-201. [ Google Scholar]
- Pishtiwan AH, Khadija KM. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli isolated from thalassemia patients in Erbil, Iraq. Mediterr J Hematol Infect Dis 2019; 11(1):e2019041. doi: 10.4084/mjhid.2019.041 [Crossref] [ Google Scholar]
- Abdul Raouf FE, Benyagoub E, Alkhudhairy MK, Akrami S, Saki M. Extended-spectrum β-lactamases among Klebsiella pneumoniae from Iraqi patients with community-acquired pneumonia. Rev Assoc Med Bras (1992) 2022; 68(6):833-7. doi: 10.1590/1806-9282.20220222 [Crossref] [ Google Scholar]
- Saisi H, Makobe C, Kangongo M, Kariuki S. Prevalence of CTX-M, SHV, TEM and OXA genes among extended-spectrum beta-lactamase producing Klebsiella pneumoniae from Mukuru Slum, Kenya. Adv Microbiol 2019; 9(10):853-62. [ Google Scholar]
- Ben-Ami R, Rodríguez-Baño J, Arslan H, Pitout JD, Quentin C, Calbo ES. A multinational survey of risk factors for infection with extended-spectrum beta-lactamase-producing Enterobacteriaceae in nonhospitalized patients. Clin Infect Dis 2009; 49(5):682-90. doi: 10.1086/604713 [Crossref] [ Google Scholar]
- Patil S, Chen X, Wen F. Exploring the phenotype and genotype of multi-drug resistant Klebsiella pneumoniae harbouring blaCTX-M group extended-spectrum β-lactamases recovered from paediatric clinical cases in Shenzhen, China. Ann Clin Microbiol Antimicrob 2019; 18(1):32. doi: 10.1186/s12941-019-0331-z [Crossref] [ Google Scholar]
- Alibi S, Ferjani A, Boukadida J. Molecular characterization of extended spectrum β-lactamases produced by Klebsiella pneumoniae clinical strains from a Tunisian hospital. Med Mal Infect 2015; 45(4):139-43. doi: 10.1016/j.medmal.2015.01.010 [Crossref] [ Google Scholar]
- Mshana SE, Hain T, Domann E, Lyamuya EF, Chakraborty T, Imirzalioglu C. Predominance of Klebsiella pneumoniae ST14 carrying CTX-M-15 causing neonatal sepsis in Tanzania. BMC Infect Dis 2013; 13:466. doi: 10.1186/1471-2334-13-466 [Crossref] [ Google Scholar]
- Livermore DM. Antibiotic resistance during and beyond COVID-19. JAC Antimicrob Resist 2021; 3(Suppl 1):i5-16. doi: 10.1093/jacamr/dlab052 [Crossref] [ Google Scholar]
- Delgado-Valverde M, Sojo-Dorado J, Pascual Á, Rodríguez-Baño J. Clinical management of infections caused by multidrug-resistant Enterobacteriaceae. Ther Adv Infect Dis 2013; 1(2):49-69. doi: 10.1177/2049936113476284 [Crossref] [ Google Scholar]
- Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev 2009; 22(1):161-82, Table of Contents. doi: 10.1128/cmr.00036-08 [Crossref] [ Google Scholar]
- Jørgensen RL, Nielsen JB, Friis-Møller A, Fjeldsøe-Nielsen H, Schønning K. Prevalence and molecular characterization of clinical isolates of Escherichia coli expressing an AmpC phenotype. J Antimicrob Chemother 2010; 65(3):460-4. doi: 10.1093/jac/dkp484 [Crossref] [ Google Scholar]
- Suwantarat N, Logan LK, Carroll KC, Bonomo RA, Simner PJ, Rudin SD. The prevalence and molecular epidemiology of multidrug-resistant Enterobacteriaceae colonization in a pediatric intensive care unit. Infect Control Hosp Epidemiol 2016; 37(5):535-43. doi: 10.1017/ice.2016.16 [Crossref] [ Google Scholar]
- Rohde AM, Zweigner J, Wiese-Posselt M, Schwab F, Behnke M, Kola A. Prevalence of third-generation cephalosporin-resistant Enterobacterales colonization on hospital admission and ESBL genotype-specific risk factors: a cross-sectional study in six German university hospitals. J Antimicrob Chemother 2020; 75(6):1631-8. doi: 10.1093/jac/dkaa052 [Crossref] [ Google Scholar]
- Tamma PD, Sharara SL, Pana ZD, Amoah J, Fisher SL, Tekle T. Molecular epidemiology of ceftriaxone non-susceptible Enterobacterales isolates in an academic medical center in the United States. Open Forum Infect Dis 2019; 6(8):ofz353. doi: 10.1093/ofid/ofz353 [Crossref] [ Google Scholar]
- Sadeghi MR, Ghotaslou R, Akhi MT, Asgharzadeh M, Hasani A. Molecular characterization of extended-spectrum β-lactamase, plasmid-mediated AmpC cephalosporinase and carbapenemase genes among Enterobacteriaceae isolates in five medical centres of East and West Azerbaijan, Iran. J Med Microbiol 2016; 65(11):1322-31. doi: 10.1099/jmm.0.000356 [Crossref] [ Google Scholar]
- Hou XH, Song XY, Ma XB, Zhang SY, Zhang JQ. Molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates. Braz J Microbiol 2015; 46(3):759-68. doi: 10.1590/s1517-838246320140138 [Crossref] [ Google Scholar]
- Aryal SC, Upreti MK, Sah AK, Ansari M, Nepal K, Dhungel B. Plasmid-mediated AmpC β-lactamase CITM and DHAM genes among gram-negative clinical isolates. Infect Drug Resist 2020; 13:4249-61. doi: 10.2147/idr.S284751 [Crossref] [ Google Scholar]