Avicenna Journal of Clinical Microbiology and Infection. 11(2):82-86.
doi: 10.34172/ajcmi.3550
Original Article
A Study of Antimicrobial Resistance Rates in Clinical Burn Center Isolates of Pseudomonas aeruginosa in the South of Iran
Elham Jafarzadeh 1  , Masoud Kargar 2, 3, 4, *
, Masoud Kargar 2, 3, 4, *  , Susan Amiri 5
, Susan Amiri 5
Author information:
1Department of Laboratory Sciences, Taleghani Hospital, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
2Department of Medical Laboratory Sciences, School of Allied Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
3Thalassemia and Hemoglobinopathy Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
4Student Research Committee, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
5Department of Biotechnology, Azad University, Ahvaz, Iran
Abstract
Background: This research aimed to examine the frequency of resistance to antimicrobial agents and their variations in Pseudomonas aeruginosa clinical isolates, which are accountable for invasive infections in the southern part of Iran, from 2018 to 2022.
Methods: A retrospective study was conducted, involving the gathering of microbiological data from Taleghani Burn Hospital from 2018 to 2022. The primary variables under scrutiny were department, sample origin, the antiprogram system (e.g., disc diffusion and strip methods, and clinical laboratory standards), and the rate or percentage of resistant isolates investigated by antimicrobial susceptibility testing. The interpretation criteria employed for the study were those of the Clinical Laboratory Standards Institute (CLSI). And the percentage of resistant isolates was also taken into consideration.
Results: The disc diffusion and strip method is the most commonly used approach for antimicrobial susceptibility testing. According to the CLSI, resistance rates ranged from 3.64% (colistin) to 77.38% (amikacin). The rates of antimicrobial resistance remained relatively constant over time in 2018-2022. Approximately 67.62% of isolates were multidrug resistant, and the remaining 9.46% were extensively drug-resistant. Wound and urine isolates demonstrated higher resistance, except for amikacin and piperacillin, than those from blood culture and biopsy.
Conclusion: Antimicrobial resistance is widely prevalent in P. aeruginosa, a common bacterium in southern Iran. The findings revealed the highest resistance rates for commonly used antibiotics such as amikacin, piperacillin, ceftazidime, and meropenem. However, colistin and nitrofurantoin are more effective against this bacterium. The wound and urine isolates represented the highest resistance rates, indicating the need for prompt and appropriate treatment. Interestingly, the resistance rates for most antibiotics remained relatively stable during the study period, emphasizing the need to develop alternative treatments for P. aeruginosa infections.
Keywords: Antibiotic resistance, Burn, Pseudomonas aeruginosa, Infection, Microbiology
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Jafarzadeh E, Kargar M, Amiri S. A study of antimicrobial resistance rates in clinical burn center isolates of pseudomonas aeruginosa in the south of Iran. Avicenna J Clin Microbiol Infect. 2024; 11(2):82-86. doi:10.34172/ajcmi.3550
Introduction
Hospital-acquired infections are a primary concern for public health, with significant social and economic impacts worldwide (1,2). Pseudomonas aeruginosa is one of the many pathogens that can lead to infections. It is considered an opportunistic pathogen that poses a severe threat, especially in cases of invasive infections, as such infections tend to have high rates of morbidity and mortality (3). What makes this microorganism a formidable foe is its innate ability to resist numerous antimicrobial treatments and develop resistance to broad-spectrum medications such as carbapenems and polymyxins (4-7).
In cases where non-multidrug-resistant (non-MDR) P. aeruginosa causes infections, there are various therapeutic options available. Some of the commonly used options are beta-lactams such as ceftazidime, cefepime, imipenem, piperacillin/tazobactam, meropenem, and aztreonam, as well as fluoroquinolones and aminoglycosides. However, the treatment of infections caused by MDR P. aeruginosa can be quite challenging. In such situations, only a limited number of antimicrobials may be available, including colistin, which has potential toxicity issues. Newer antimicrobials such as ceftolozane/tazobactam and ceftazidime/avibactam, which have less clinical experience, may be used alternatively (8-10). To select the most effective antimicrobial treatment for a given infection caused by P. aeruginosa, it is essential to have a clear understanding of the local susceptibility and resistance rates. These rates can vary depending on different factors, such as the geographical region, local resistance patterns, and microbial factors such as the specific clone and resistance mechanisms involved. Unfortunately, the available data on antimicrobial resistance rates in P. aeruginosa isolates causing invasive infections in Iran are limited, making it challenging to develop effective treatment strategies for these infections (11-14).
Therefore, this research not only provides valuable insights but also has practical implications. First, it aims to determine the prevalence of antimicrobial resistance in P. aeruginosa isolates causing invasive infections in a burn center hospital in southern Iran, categorized by sample type. Second, it seeks to investigate how these antimicrobial resistance rates have changed from 2018 to 2022. These findings can directly impact patient outcomes by enabling the development of targeted and efficient treatment strategies, making this research highly relevant and applicable in the field of healthcare.
Methods
This study has provided a detailed retrospective analysis of the demographic and microbiological data collected from 2018 to 2022 in the Taleghani Hospital Burn Center in Iran. The data include various parameters such as age, gender, and underlying health conditions of the patients. The study has focused on the first isolates of each infection type per year, which includes bacteremia, wound infections, nosocomial urinary tract infections, and biopsies. Only the first 3 isolates were considered for patients with more than one isolate obtained per type of infection and year. The study variables were carefully selected, including the department, sample origin, the antiprogram system (e.g., disc diffusion and strip methods and clinical laboratory standards), and the rate or percentage of resistant isolates. The bacterial identification and susceptibility tests were conducted for specified antimicrobials at each site. The analyzed antimicrobials included piperacillin, ceftazidime, trimethoprim-sulfamethoxazole, imipenem, meropenem, nitrofurantoin, gentamicin, amikacin, ciprofloxacin, and colistin. The corresponding clinical category for each antimicrobial agent was determined based on the Clinical Laboratory Standards Institute (CLSI) 2020 (15). The parameters were statistically analyzed using SPSS for Windows statistical software (version 20; SPSS, Inc., Chicago, IL, USA). Categorical variables were evaluated using either the chi-square test or Fisher’s exact test. A P value of less than 0.05 was considered statistically significant.
Results
Out of the 896 data points analyzed, the origin by type of unit or department was as follows:
About 20% were men, 5.1% were children, 66.3% were from the ICU, and 8.6% were women. The isolates were obtained from wounds (8.81%), urine (13.28%), blood culture (14.73%), biopsy (63.16%), and other samples (0.02%).
The results of disc diffusion and strip methods showed that the proportion of resistant isolates varied between 3.64% (colistin) and 77.38% (amikacin). Only the overall percentage of resistant isolates from 2018 to 2022 demonstrated statistically significant differences among all tested antimicrobials (Table 1).
Table 1.
Distribution of Resistance Rates per Year and Overall Resistance Rates Based on the Applied Breakpoint (CLSI)
|
|
Percentage Resistant (R) Isolates by
|
|
Antimicrobial
|
Year (%R)
|
Breakpoint
|
|
2018
|
2019
|
2020
|
2021
|
2022
|
CLSI
%Ra
|
| Nitrofurantoin |
35.6 |
30 |
33.3 |
32.3 |
30.8 |
32.4 |
| Colistin |
3.3 |
3.5 |
4 |
3.6 |
3.8 |
3.64 |
| Ceftazidime |
72.2 |
71.4 |
73.1 |
72.5 |
74.1 |
72.66 |
| Ciprofloxacin |
67.7 |
71.6 |
70.2 |
70.3 |
70 |
69.69 |
| Trimethoprim |
61.7 |
60 |
61.4 |
61.5 |
60.7 |
61.06 |
| Amikacin |
76.2 |
78.3 |
76.9 |
76.9 |
78.6 |
77.38 |
| Gentamicin |
67.5 |
69.6 |
69.2 |
69.8 |
67.5 |
68.72 |
| Imipenem |
67.3 |
67.8 |
67.1 |
67.6 |
67.5 |
67.46 |
| Meropenem |
72.2 |
69.6 |
71.8 |
72.2 |
73.7 |
71.9 |
| Piperacillin |
72.4 |
73.5 |
72 |
72.6 |
73.4 |
72.78 |
Note. CLSI: Clinical laboratory standards institute; a Resistance isolates.
Moreover, the resistance rates were consistent primarily from 2018 to 2022 (Table 1). However, there were a few exceptions. For example, ciprofloxacin and ceftazidime rates decreased from 70.3% in 2021 to 70% in 2022 (P = 0.001) and from 73.1% in 2020 to 72.5% in 2021 (P = 0.03), respectively. In addition, imipenem and colistin rates decreased from 67.6% in 2021 to 67.5% in 2022 (P = 0.001) and from 4.0% in 2020 to 3.6% in 2021 (P = 0.006), respectively.
No cases of pan-resistant isolates were identified based on the criteria by Magiorakos et al (16). However, 67.62% of the isolates were found to be MDR, while 9.46% were classified as extensively drug-resistant (XDR). Figure 1 depicts a modest rise in the percentage of MDR isolates from 50% in 2018 to 77.9% in 2022. On the other hand, there has been a decline in XDR isolates from 24.2% in 2018 to 1.8% in 2022. The microbial isolates extracted from urine and wound samples exhibited higher resistance to the tested antimicrobial agents, especially amikacin, ceftazidime, and imipenem (up to 40%). Meanwhile, the microorganisms obtained from blood and biopsy samples were less resistant to the same antimicrobial agents (as per Table 2).
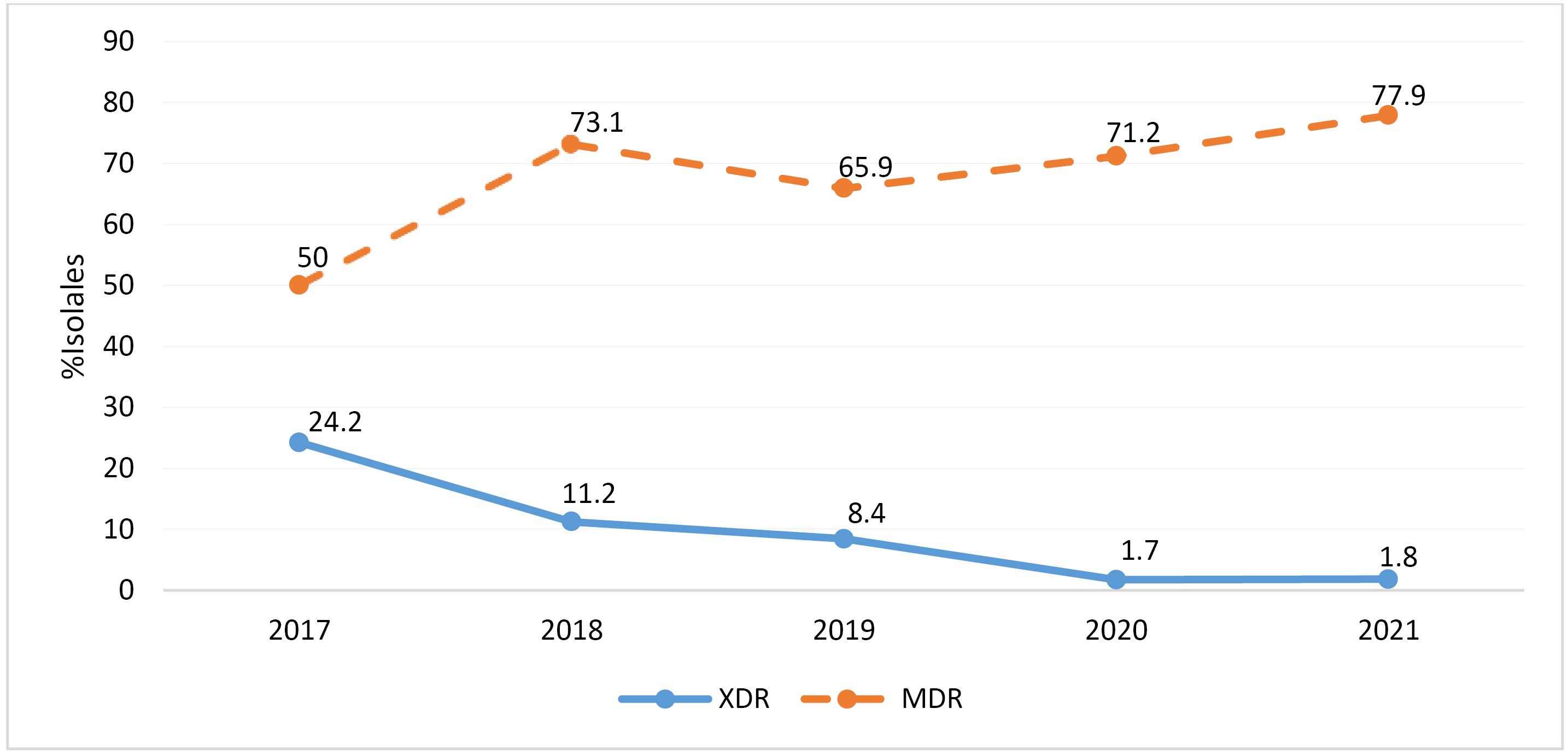
Figure 1.
Annual Distribution of the Percentages of MDR and XDR P. aeruginosa Isolates. Note. P. aeruginosa: Pseudomonas aeruginosa; MDR: Multidrug-resistant; XDR: Extensively drug-resistant
.
Annual Distribution of the Percentages of MDR and XDR P. aeruginosa Isolates. Note. P. aeruginosa: Pseudomonas aeruginosa; MDR: Multidrug-resistant; XDR: Extensively drug-resistant
Table 2.
Distribution of Antimicrobial Resistance Rates by Sample Type
|
Antimicrobial
|
Percentage Resistant Isolates (%R) by Type of Sample
|
|
Biopsya
(63.16%)
|
Blood Culturea
(14.73%)
|
Urine Culturea
(13.28%)
|
Wounda
(8.81%)
|
| Nitrofurantoin |
0 |
0 |
31.1 |
0 |
| Colistin |
1.9 |
3.0 |
5.0 |
6.3 |
| Ceftazidime |
36.7 |
40.2 |
38.7 |
43.0 |
| Ciprofloxacin |
21.7 |
25.0 |
29.4 |
36.7 |
| Trimethoprim |
12.0 |
6.8 |
11.8 |
12.7 |
| Amikacin |
47.5 |
53.0 |
43.7 |
48.1 |
| Gentamicin |
18.9 |
8.3 |
37.0 |
26.6 |
| Imipenem |
40.1 |
39.4 |
40.3 |
40.5 |
| Meropenem |
21.2 |
20.5 |
29.4 |
22.8 |
| Piperacillin |
35.1 |
40.2 |
26.9 |
26.6 |
Note. a Overall, 0.02% of other samples are not included.
Discussion
According to the results, P. aeruginosa was the hospitals’ most commonly acquired pathogen. Its resistance to antimicrobial drugs has significantly increased in recent years, albeit with varying rates (14). Knowing the local rates of antimicrobial resistance and their trends over time is necessary for designing appropriate therapeutic strategies, particularly those that are empirical, as this justifies their requirement.
The findings of this research showed that the prevalence of antimicrobial resistance in P. aeruginosa at Taleghani Hospital in the southern region of Iran, Ahvaz, was relatively low compared to those reported in other studies (14,17,18), and resistance rates varied according to those of the CLSI.
The resistance rate was the highest with amikacin (77.38%), irrespective of the applied CLSI. Colistin was found to be the most active antibiotic, with a resistance rate of less than 10% (19).
These findings are in line with previous research outcomes, including those of a clinical burn care study in Iran that examined isolates collected between May 2012 and November 2014. In that study, the highest levels of resistance were detected with ciprofloxacin (93.3%), imipenem (60%), gentamicin (60%), and amikacin (66.6%). One of the most effective antimicrobials was trimethoprim (33.3%) (14). Similar or almost identical trends were observed in recent studies conducted in Iran. The results of a study performed in Tehran between September 2005 and October 2007 revealed that the highest resistance rates were associated with ciprofloxacin (72%), gentamicin (80%), and amikacin (73%) (20). These findings corroborate those of research conducted in Isfahan using 2016 samples. The research demonstrated that the highest resistance rates were related to ciprofloxacin, imipenem, meropenem, amikacin, and ceftazidime, all of which were over 90%. Additionally, the resistance rates to piperacillin were 70.7%. The study identified colistin as the most effective antibiotic (21).In 2015, in a study conducted in Ardabil, researchers evaluated 94 strains of P. aeruginosa and reported that out of 83 isolates, the highest resistance rates were observed in imipenem (75.5%), gentamicin (86%), ciprofloxacin (87%), amikacin (85%), and piperacillin (82%). On the other hand, colistin was found to be the most effective antibiotic with a resistance rate of 0% (22).
Based on previous analysis (2018-2022), a thorough examination of antimicrobials revealed little to no significant developments or discernible patterns in the resistance rates. However, the resistance rate of piperacillin showed a noteworthy change; it was reduced by half (73.5–72%) in 2019 compared to 2020. These findings suggest that there should be significant changes in the hospitals in southern Iran regarding the distribution of resistant determinants or resistant clones. Observations of this trend date back to the early 2000s, as evidenced by the results of prior research performed in Iran (14,20).
No pan-resistant isolates were identified in our study. However, the results indicated a relatively high prevalence rate of MDR isolates (67.62%) and a low prevalence rate of XDR isolates (9.46%). These combined resistance percentages are comparable to those found in some countries. For example, in India, the prevalence of pan-resistant isolates was 4%, while MDR and XDR isolates were found to have a prevalence of 50% and 2.3%, respectively (23).
The microbial isolates extracted from urine and wound samples represented comparatively higher resistance to the tested antimicrobials, particularly amikacin and piperacillin, than those isolated from other sources such as biopsy or blood. This outcome is consistent with that of previous research and may be linked to the escalated usage of antimicrobials for treating P. aeruginosa-induced wound and urine infections (18). This study’s findings are subject to certain limitations, primarily due to the lack of verification by a reference laboratory of the reported data on P. aeruginosa susceptibility. Nonetheless, this study’s comprehensive scope, encompassing numerous participating sites, types of samples, and years, confers a reasonably accurate understanding of P. aeruginosa susceptibility in the southern region of our country. In the absence of national standardized databases, such studies prove essential in obtaining such information. A further limitation of this work pertains to the absence of molecular epidemiology studies, such as the pulsed-field gel electrophoresis (PFGE) or multi-locus sequence typing (MLST), which would have enabled the determination of whether the observed prevalence of resistance is influenced by certain endemic situations, such as resistant clones or horizontal resistance determinants, in the participating sites (13,14,18). Certain factors, including the dosage of antimicrobials and the infection’s focus, can affect isolates’ susceptibility. This modification will result in a substantial reduction in resistance rates for several antimicrobials (24).
Conclusion
Antimicrobial resistance is widely prevalent in P. aeruginosa, a bacterium commonly present in southern Iran. The findings revealed the highest resistance rates for commonly used antibiotics such as amikacin, piperacillin, ceftazidime, and meropenem. However, colistin and nitrofurantoin were more effective against this bacterium. The wound and urine isolates demonstrated the highest resistance rates, indicating the need for prompt and appropriate treatment. Interestingly, the resistance rates for most antibiotics remained relatively stable during the study period, highlighting the need for developing alternative treatments for P. aeruginosa-related infections.
Authors’ Contribution
Conceptualization: Elham Jafarzadeh.
Methodology: Masoud Kargar.
Writing–original draft: Elham Jafarzadeh.
Writing–review & editing: Masoud Kargar, Susan Amiri.
Competing Interests
All authors declare no conflict of interests.
Ethical Approval
Not applicable.
Funding
The authors received no funding support.
References
- Olaechea PM, Insausti J, Blanco A, Luque P. [Epidemiology and impact of nosocomial infections]. Med Intensiva 2010; 34(4):256-67. doi: 10.1016/j.medin.2009.11.013.[Spanish] [Crossref] [ Google Scholar]
- Mo Y, Low I, Tambyah SK, Tambyah PA. The socio-economic impact of multidrug-resistant nosocomial infections: a qualitative study. J Hosp Infect 2019; 102(4):454-60. doi: 10.1016/j.jhin.2018.08.013 [Crossref] [ Google Scholar]
- de Matos EC, Andriolo RB, Rodrigues YC, de Lima PD, do Rosário Souza Carneiro IC, Lima KV. Mortality in patients with multidrug-resistant Pseudomonas aeruginosa infections: a meta-analysis. Rev Soc Bras Med Trop 2018; 51(4):415-20. doi: 10.1590/0037-8682-0506-2017 [Crossref] [ Google Scholar]
- Sedighi M, Ghasemian Safaie H, Vaez H, Moghoofeie M, Hadifar S, Oryan G, et al. Detection of blaSPM-1 metallo-β-lactamase gene in Imipenem-resistant Pseudomonas aeruginosa strains isolated from hospitalized patients in Isfahan hospitals. Journal of Microbial Biology 2015;4(13):161-70. [Persian].
- Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents 2015; 45(6):568-85. doi: 10.1016/j.ijantimicag.2015.03.001 [Crossref] [ Google Scholar]
- López-Causapé C, Cabot G, Del Barrio-Tofiño E, Oliver A. The versatile mutational resistome of Pseudomonas aeruginosa. Front Microbiol 2018; 9:685. doi: 10.3389/fmicb.2018.00685 [Crossref] [ Google Scholar]
- Fernández-Cuenca F, Martínez-Martínez L, Pascual Á. Evolution of the antimicrobial resistance rates in clinical isolates of Pseudomonas aeruginosa causing invasive infections in the south of Spain. Enferm Infecc Microbiol Clin (Engl Ed) 2020; 38(4):150-4. doi: 10.1016/j.eimc.2019.06.009 [Crossref] [ Google Scholar]
- Bassetti M, Vena A, Russo A, Croxatto A, Calandra T, Guery B. Rational approach in the management of Pseudomonas aeruginosa infections. Curr Opin Infect Dis 2018; 31(6):578-86. doi: 10.1097/qco.0000000000000505 [Crossref] [ Google Scholar]
- Monogue ML, Nicolau DP. Antibacterial activity of ceftolozane/tazobactam alone and in combination with other antimicrobial agents against MDR Pseudomonas aeruginosa. J Antimicrob Chemother 2018; 73(4):942-52. doi: 10.1093/jac/dkx483 [Crossref] [ Google Scholar]
- Wright H, Bonomo RA, Paterson DL. New agents for the treatment of infections with gram-negative bacteria: restoring the miracle or false dawn?. Clin Microbiol Infect 2017; 23(10):704-12. doi: 10.1016/j.cmi.2017.09.001 [Crossref] [ Google Scholar]
- Kalantar E, Torabi V, Salimizand H, Soheili F, Ramezanzadeh R. Incidence and susceptibility pattern of metallo-beta-lactamase producers among Pseudomonas aeruginosa isolated from burn patients at Kurdistan province. Jundishapur J Microbiol 2012; 5(3):507-10. doi: 10.5812/jjm.3664 [Crossref] [ Google Scholar]
- Ekrami A, Abbasi Montazeri E, Kaydani GA, Shokoohizadeh L. Methicillin resistant staphylococci: prevalence and susceptibility patterns in a burn center in Ahvaz from 2013-2014. Iran J Microbiol 2015; 7(4):208-13. [ Google Scholar]
- Hassanzadeh P, Motamedifar M, Hadi N. Prevalent bacterial infections in intensive care units of Shiraz University of Medical Sciences Teaching Hospitals, Shiraz, Iran. Jpn J Infect Dis 2009; 62(4):249-53. [ Google Scholar]
- Shams S, Ghorbanalizadgan M, Minoii S, Shojaei S. Evaluation of bacterial infection of burn wounds in a burn center, Qom, Iran. Arch Hyg Sci 2018; 7(4):282-7. doi: 10.29252/ArchHygSci.7.4.282 [Crossref] [ Google Scholar]
- Weinstein MP, Lewis JS 2nd. The clinical and laboratory standards institute subcommittee on antimicrobial susceptibility testing: background, organization, functions, and processes. J Clin Microbiol 2020; 58(3):e01864-19. doi: 10.1128/jcm.01864-19 [Crossref] [ Google Scholar]
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18(3):268-81. doi: 10.1111/j.1469-0691.2011.03570.x [Crossref] [ Google Scholar]
- Afkhamzadeh A, Majidi F, Ahmadi C. Risk factors for nosocomial infections among burn patients hospitalized in Tohid hospital, Sanandaj, Kurdistan Iran. Med J Mashhad Univ Med Sci 2016; 59(4):225-32. doi: 10.22038/mjms.2016.8482.[Persian] [Crossref] [ Google Scholar]
- Beheshti S, Zia M. Bacteriology of burns and antibiogram in an Iranian burn care center. Afr J Pharm Pharmacol 2011; 5(4):538-41. doi: 10.5897/ajpp10.375 [Crossref] [ Google Scholar]
- Mastoraki A, Douka E, Kriaras I, Stravopodis G, Manoli H, Geroulanos S. Pseudomonas aeruginosa susceptible only to colistin in intensive care unit patients. Surg Infect (Larchmt) 2008; 9(2):153-60. doi: 10.1089/sur.2007.004 [Crossref] [ Google Scholar]
- Bojary Nasrabadi MR, Hajia M. Multidrug-resistant Pseudomonas aeruginosa strains in Tehran reference burn hospital, Tehran, Iran. Afr J Microbiol Res 2012; 6(7):1393-6. doi: 10.5897/ajmr11.1048 [Crossref] [ Google Scholar]
- Radan M, Moniri R, Khorshidi A, Gilasi H, Norouzi Z, Beigi F. Emerging carbapenem-resistant Pseudomonas aeruginosa isolates carrying blaIMP among burn patients in Isfahan, Iran. Arch Trauma Res 2016; 5(3):e33664. doi: 10.5812/atr.33664 [Crossref] [ Google Scholar]
- Adabi M, Talebi Taher M, Arbabi L, Afshar M, Fathizadeh S, Minaeian S, et al. Determination of antibiotic resistance pattern of Pseudomonas aeruginosa strains isolated from patients with burn wounds. J Ardabil Univ Med Sci 2015;15(1):66-74. [Persian].
- Gill JS, Arora S, Khanna SP, Kumar KH. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level intensive care unit. J Glob Infect Dis 2016; 8(4):155-9. doi: 10.4103/0974-777x.192962 [Crossref] [ Google Scholar]
- Kahlmeter G. Redefining Susceptibility Testing Categories S, I and R. Växjö, Sweden: European Committee on Antimicrobial Susceptibility Testing (EUCAST); 2019.