Avicenna Journal of Clinical Microbiology and Infection. 11(4):156-162.
doi: 10.34172/ajcmi.3558
Original Article
Investigating antimicrobial resistance in Salmonella strains isolated from food in Syria
Mazen Safi 1, *  , Bassam Al balaa 1, Samah Qasem 1, Laila Al Hallab 1, Ayman Al-mariri 1
, Bassam Al balaa 1, Samah Qasem 1, Laila Al Hallab 1, Ayman Al-mariri 1
Author information:
1Department of Molecular Biology and Biotechnology, Atomic Energy Commission, Damascus, Syria
Abstract
Background: The emergence of multiple drug resistance in Salmonella typhi and Salmonella paratyphi poses a significant challenge, necessitating the development of effective treatments to combat these bacteria and reduce infection rates. This in vitro study aimed to evaluate the minimum inhibitory concentration (MIC) of various antibiotics against S. typhi and S. paratyphi.
Methods: Overall, 116 samples were collected from diverse markets in Syria. Molecular techniques, including polymerase chain reaction, were utilized to identify the bacterial genus. Antimicrobial susceptibility testing, employing the disk diffusion method and MIC determination, was conducted to assess the effectiveness of various antibiotics.
Results: Among the isolates, 46 were identified, consisting of 23 S. typhi and 23 S. paratyphi strains. Resistance to nalidixic acid was observed in 9 out of 23 S. typhi and 11 out of 23 S. paratyphi isolates. Notably, these nalidixic acid-resistant strains exhibited elevated MIC50 values for other fluoroquinolones. Furthermore, most of these resistant isolates, specifically 8 out of 9 S. typhi and 11 out of 11 S. paratyphi, displayed complete resistance to ciprofloxacin (MIC50≥2 μg/L).
Conclusion: Based on our findings, only gentamicin, third-generation cephalosporins, and some fluoroquinolones demonstrated efficacy effects against S. typhi and S. paratyphi isolates in this study.
Keywords: Salmonella typhi, Salmonella paratyphi, Enteric fever, Fluoroquinolones, Drug therapy, Drug resistance
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Safi M, Al balaa B, Qasem S, Al Hallab L, Al-mariri A. Investigating antimicrobial resistance in Salmonella strains isolated from food in Syria. Avicenna J Clin Microbiol Infect. 2024; 11(4):156-162. doi:10.34172/ajcmi.3558
Introduction
Typhoid fever, a significant public health concern, disproportionately impacts regions with inadequate access to clean water sources and proper sanitation facilities. This infectious disease affects approximately 11-20 million individuals globally, resulting in approximately 128 000 to 161 000 deaths annually (1). In developed countries, most cases of typhoid fever are acquired through travelling to endemic regions or among immigrants from these areas (2). According to World Health Organization reports, 2 billion people worldwide experience diarrhea, with one-third of these cases attributed to contaminated food sources (3).
Approximately 1.5 million cases of salmonellosis globally occur each year due to direct contact with animals such as dogs, ruminants, and horses (4). Salmonella, a bacterium commonly associated with animals, is prevalent in their populations and can be transmitted through their products. This pathogen causes intestinal inflammation in poultry, leading to a high mortality rate (5). In addition, Salmonella can result in the deaths of newborn calves (6). While not typically part of poultry’s natural intestinal flora, Salmonella can be acquired from the environment via live insects, rodents, and contaminated feed. Infected adult animals may not exhibit outward symptoms, and the infection can be spread through pasture, barn bedding, and milking equipment (7). Furthermore, Salmonella can also be transmitted through veterinary tools used during animal examination or from the infected animal to the veterinarians (8).
Salmonella is a significant bacterium commonly transmitted through contaminated food and is capable of causing typhoid fever (9). Meat, particularly poultry and pig meat, is a major source of Salmonella contamination (10).
The Salmonella genus is comprised of S. bongori and S. enterica species, with S. enterica further divided into six subspecies. S. typhi and S. paratyphi belong to the S. enterica subsp.enteric subspecies (11,12).
Antibiotics are crucial in the treatment of typhoid fever. Chloramphenicol was first successfully used to treat patients with typhoid fever in 1948 (13). Chloramphenicol became the preferred drug for treating typhoid fever until the 1970s, when the first outbreak of infections caused by antibiotic-resistant bacteria was reported (14). There has been a rise in resistance among S. typhi strains to chloramphenicol, as well as other drugs such as ampicillin and trimethoprim-sulfamethoxazole. The emergence of resistance to multiple antibiotics poses a significant challenge in managing typhoid fever (15,16). Fluoroquinolones have shown efficacy in treating multidrug-resistant (MDR) typhoid fever (17). Unfortunately, the prevalence of nalidixic acid-resistant Salmonella strains, which also exhibit resistance to other fluoroquinolones, has been observed in various countries (18). The detection of Salmonella isolates with reduced sensitivity to fluoroquinolones and third-generation cephalosporin, such as ceftriaxone is concerning (19,20).
Salmonella-induced diseases can impose significant stress on individuals, leading to extended periods of work disability. While the mortality rate associated with Salmonella infections is relatively low, the treatment costs are high, particularly due to the rising resistance of these bacteria to multiple antibiotics. Research into the effects of certain antibiotics on Syrian Salmonella isolates in vitro could provide insights for potential in vivo applications, aiming to alleviate symptoms and improve treatment outcomes in the future.
The current in vitro study aims to assess the minimum inhibitory concentration (MIC) of various antibiotics against S. typhi and S. paratyphi. This research is crucial for evaluating the therapeutic significance of these antibiotics in treating infections caused by these bacteria. By determining the effectiveness of different antibiotics against these specific Salmonella strains, the study can provide valuable information for guiding treatment strategies and improving patient outcomes.
Materials and Methods
Samples Collection
A total of 116 different samples were collected between March 2022 and April 2023, comprising 46 milk samples, 34 chicken meat samples, and 36 egg samples.
Isolation of Bacteria
The bacteria were isolated using lysine iron agar as a selective medium for intestinal bacteria. Subsequently, the Salmonella agar medium was utilized as a differential medium for the Salmonella genus, following previously described protocols (21).
Polymerase Chain Reaction
Utilizing the Salmonella gene database and Primer 5 software, three specific primer pairs were designed based on the nucleotide sequence of genes. The first primer pair was designed to amplify the invA gene specific to the Salmonella genus. The second primer pair targeted the prt gene responsible for encoding the enzyme involved in paratose sugar synthesis, found in both S. typhi and S. paratyphi serotypes but absent in other Salmonella serotypes. To differentiate between S. typhi and S. paratyphi, a third primer pair was designed to amplify the tyv gene specific to S. typhi, encoding the enzyme CDP tyvelose-2-epimerase (Table 1).
Table 1.
Specific Primers Used in PCR
|
Primer
|
Nucleotide Sequence
|
| P5` tyv |
5`-GAG GAA GGG AAA TGA AGC TTT T-3` |
| P3` tyv |
5`-TAG CAA ACT GTC TCC CAC CAT AC-3` |
| P5` p r t |
5`-CTT GCT ATG GAA GAC ATA ACG AAC C-3` |
| P3` pr t |
5`-CGT CTC CAT CAA AAG CTC CAT AGA-3` |
| P5` inv |
5`-GTA TTG TTG ATT AAT GAG ATC CG-3` |
| P3` inv |
5`-ATA TTA CGC ACG GAA ACA CGT T-3` |
Note. PCR: Polymerase chain reaction.
Selected isolates were prepared for multiplex PCR. The reaction mixture was prepared and worked under conditions provided in Tables 2 and 3.
Table 2.
Materials used in PCR
|
Samples
|
Volume
|
| DNA template (colony) |
5 µL |
| Buffer (10X) |
2.5 µL |
| MgSO4 (50 mM) |
1 µL |
| dNTPs (10 mM) |
1 µL |
| DNA polymerase |
0.2 µL |
| Primers 5` (25 pmole/µL) |
3 µL |
| Primers 3` (25 pmole/µL) |
3 µL |
| H2O |
9.3 µL |
| Total |
25 µL |
Note. PCR: Polymerase chain reaction; MgSO4: Magnesium sulfate; dNTP: Deoxynucleotide.
Table 3.
PCR Conditions
|
Initial Denaturation
|
95°C
|
5 Minutes
|
| Cycles |
35 |
Cycles |
| Denaturation |
94°C |
1 minute |
| Annealing |
60°C |
1 minute |
| Extension |
72°C |
1 minute |
| Final extension |
72°C |
10 minutes |
Note. PCR: Polymerase chain reaction.
Antibiotic Sensitivity Tests
Disc Diffusion Method
The disc diffusion method was employed to test antibiotic susceptibility using different antibiotic tablets (Difco) at the indicated concentrations in μg, including nalidixic acid (30), azithromycin (15), ciprofloxacin (5), norfloxacin (10), tobramycin (10), rifampicin (30), nitrofurantoin (300), imipenem (10), and gentamicin (200).
Minimum Inhibitory Concentration Method
A good broth microdilution method was utilized with 96-well plates (TPP, Switzerland) as per the prior protocol (22). The MIC50 values were determined following the recommendations of the Clinical and Laboratory Standards Institute (23). The investigated antibiotics included cefprozil (Bristol-Myers Squibb, New York, USA), ceftazidime (Sigma, St. Louis, USA), ciprofloxacin (Bayer, Istanbul, Turkey), ofloxacin (Sigma), levofloxacin (Sigma), and lomefloxacin (Sigma), sparfloxacin (Sigma). The other antibiotics were gentamicin (Medochemie Limited, Limassol, Cyprus), chloramphenicol (Pfizer, New York, USA), and minocycline (Home Sunshine Pharma, Hefei City, China).
Results
Isolation of Bacteria
Overall, 68 samples containing Salmonella (78.88%) were obtained when cultured on an iron lysine agar medium. The medium maintained its violet color, and the colonies appeared transparent with or without a dark center. Positive isolates were found in milk (27), chicken meat (12), and eggs (29).
In general, 61 positive samples containing Salmonella (70.76%) were obtained when cultured on the Salmonella agar medium. The medium turned yellow around the isolates, with the isolates appearing transparent with or without a dark center. The isolates were positive in milk (24), chicken meat (10), and eggs (27).
Polymerase Chain Reaction Analysis
Through multiplex PCR, it was determined that 23 isolates were genotyped as S. typhi, with 12 in milk, 5 in chicken, and 6 in eggs. However, 23 isolates were genotyped as S. paratyphi, with 6 in milk, 5 in chicken, and 12 in eggs. Finally, 15 isolates were genotyped as Salmonella spp., with 6 in milk and 9 in eggs.
The PCR analysis showed that the isolates belonged to different Salmonella serotypes based on the bands with specific molecular weights on the agarose gel (Figure 1).
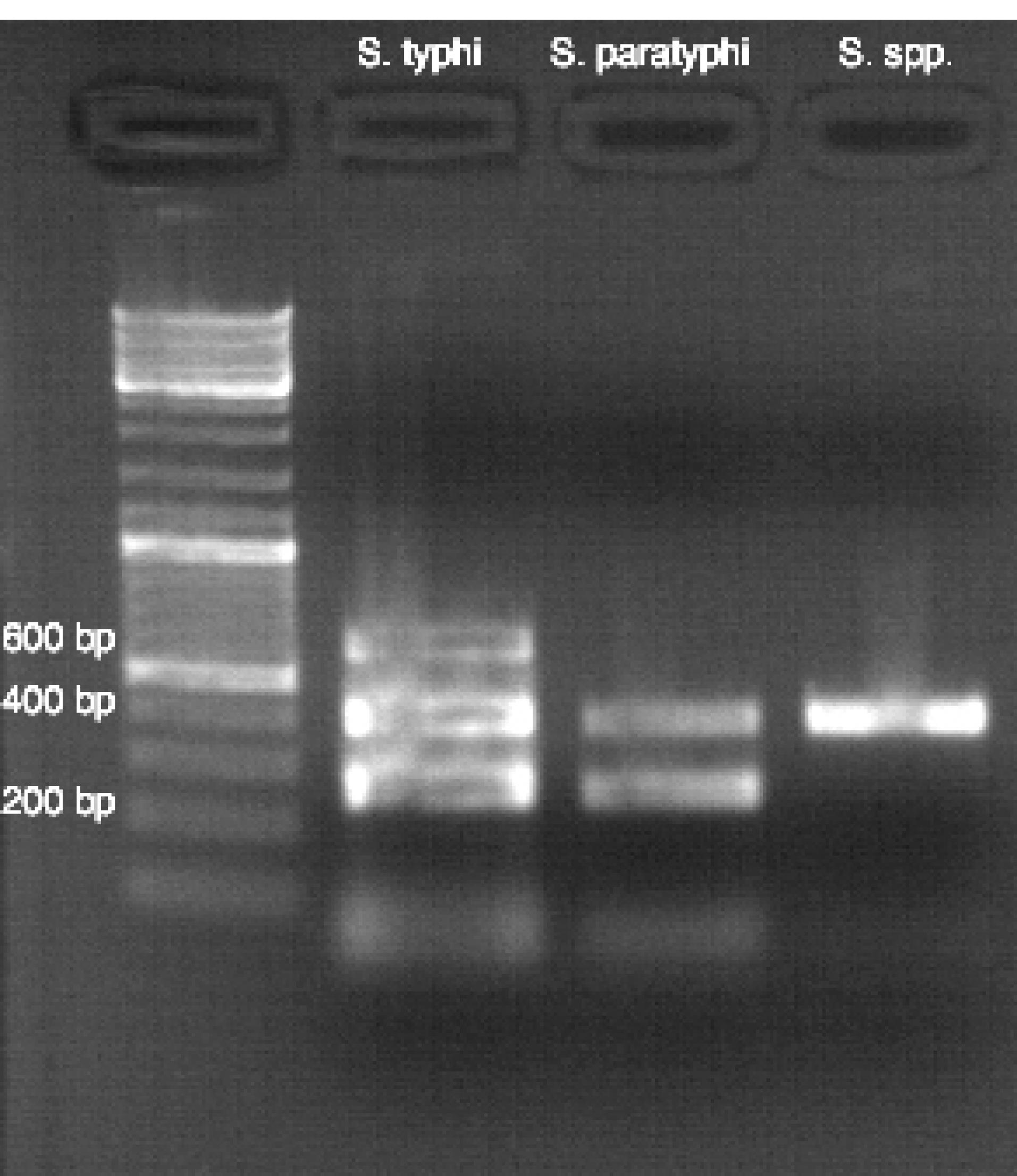
Figure 1.
Agarose Gel Electrophoresis Results by Multiplex PCR. Note. PCR: Polymerase chain reaction; MW: Standard DNA marker (GeneRulerTM 100 bp). Path 1: Negative control (water), Path 2: Salmonella typhi serotype, Path 3: S. paratyphi serotype, and Path 4: Salmonella genus
.
Agarose Gel Electrophoresis Results by Multiplex PCR. Note. PCR: Polymerase chain reaction; MW: Standard DNA marker (GeneRulerTM 100 bp). Path 1: Negative control (water), Path 2: Salmonella typhi serotype, Path 3: S. paratyphi serotype, and Path 4: Salmonella genus
Antibiotic Sensitivity Tests
Disc Diffusion Method
Based on the results (Table 4), all isolates were completely resistant to norfloxacin, nitrofurantoin, and imipenem. Tobramycin and rifampicin demonstrated weak effectiveness on 11 (24%) and 14 (30.4%) isolates, respectively. Azithromycin was the most effective antibiotic, representing susceptibility in 42 out of 46 isolates (91.3%). Gentamicin was effective in 38 out of 46 isolates (82.6%). Ciprofloxacin and nalidixic acid showed moderate effectiveness, with both antibiotics indicating susceptibility in 26 out of 46 isolates (56.5%).
Table 4.
Number of Susceptible Isolates of Salmonella typhi and Salmonella paratyphi Seriotypes Using Disc Diffusion Method
|
Isolate Resource
|
Isolate Type and Number
|
Susceptible Isolates (N)
|
|
Na.ac
|
Azit.
|
Cip.
|
Norf.
|
Tobr.
|
Rifa.
|
Nitr.
|
Imip.
|
Gent.
|
| Milk |
Typi
|
12 |
8 |
10 |
7 |
0 |
3 |
5 |
0 |
0 |
10 |
|
Paratyphi
|
6 |
4 |
5 |
4 |
0 |
2 |
3 |
0 |
0 |
5 |
| Chicken meat |
Typi
|
5 |
4 |
5 |
4 |
0 |
1 |
1 |
0 |
0 |
4 |
|
Paratyphi
|
5 |
4 |
5 |
4 |
0 |
2 |
2 |
0 |
0 |
4 |
| Eggs |
Typi
|
6 |
2 |
6 |
2 |
0 |
1 |
1 |
0 |
0 |
5 |
|
Paratyphi
|
12 |
4 |
11 |
5 |
0 |
2 |
2 |
0 |
0 |
10 |
Note. Na.ac: Nalidixic acid; Azit.: Azithromycin; Cip.: Ciprofloxacin; Norf.: Norfloxacin; Tobr.: Tobramycin; Rifa.: Rifampicin; Nitr.: Nitrofurantoin; Imip.: Imipenem; Gent.: Gentamicin.
Antibiotics Minimum Inhibitory Concentration Method
Minimum Inhibitory Concentrations Against Salmonella typhi Isolates Susceptible to Nalidixic Acid
The findings (Table 5) demonstrated that chloramphenicol (MIC50mean = 0.2 μg/mL), gentamicin (MIC50mean = 0.38 μg/mL), and Ceftazidime (MIC50mean = 0.45 μg/mL) were the most effective antibiotics against 14 S. typhi isolates susceptible to nalidixic acid. All isolates were sensitive to these antibiotics. In contrast, all isolates were completely resistant to cefprozil and minocycline. In addition, most fluoroquinolones showed high effectiveness against these isolates, except for sparfloxacin, which had moderate effectiveness (MIC50mean = 1.38 μg/mL). The MIC50mean was 0.37 μg/mL, 0.49 μg/mL, 0.28 μg/mL, and 0.31 μg/mL for lomefloxacin, levofloxacin, ciprofloxacin, and ofloxacin, respectively.
Table 5.
MIC50 in Salmonella typhi Isolates Susceptible to Nalidixic Acid
|
Mean of MIC50
|
Antibiotics
|
Concentrations of Antibiotics and Number of Isolates Susceptible to Each Antibiotic at Each Concentration
|
|
0.125
|
0.25
|
0.5
|
1
|
2
|
4
|
8
|
16
|
| 12 |
Cefprozil |
0 |
0 |
0 |
0 |
0 |
2 |
4 |
8 |
| 0.45 |
Ceftazidime |
4 |
5 |
1 |
4 |
0 |
0 |
0 |
0 |
| 0.2 |
Chloramphenicol |
6 |
8 |
0 |
0 |
0 |
0 |
0 |
0 |
| 0.38 |
Gentamicin |
2 |
8 |
2 |
2 |
0 |
0 |
0 |
0 |
| 6.2 |
Minocycline |
0 |
0 |
0 |
1 |
1 |
3 |
5 |
4 |
| 0.37 |
Lomefloxacin |
1 |
8 |
4 |
1 |
0 |
0 |
0 |
0 |
| 0.49 |
Levofloxacin |
1 |
3 |
8 |
2 |
0 |
0 |
0 |
0 |
| 0.28 |
Ciprofloxacin |
5 |
5 |
4 |
0 |
0 |
0 |
0 |
0 |
| 1.38 |
Sparfloxacin |
2 |
2 |
3 |
2 |
4 |
1 |
0 |
0 |
| 0.31 |
Ofloxacin |
5 |
5 |
3 |
1 |
0 |
0 |
0 |
0 |
Note. MIC: Minimum inhibitory concentration.
Minimum Inhibitory Concentrations Against Salmonella typhi Isolates Resistant to Nalidixic Acid
The results (Table 6) revealed that gentamicin (MIC50mean = 0.64 μg/mL) was the most effective antibiotic against 9 S. typhi isolates resistant to nalidixic acid. All isolates were sensitive to this antibiotic. All isolates were completely resistant to cefprozil and minocycline, whereas lomefloxacin and levofloxacin showed good efficacy against these isolates (MIC50mean = 1.22 and MIC50mean = 1.28 μg/mL, respectively), and only 6 isolates were susceptible to both antibiotics (67%). On the other hand, 5 isolates were sensitive to chloramphenicol and ceftazidime (56%), 2 isolates were sensitive to sparfloxacin and ofloxacin (22%), and only one isolate was sensitive to ciprofloxacin (11%).
Table 6.
MIC50 in Salmonella typhi Isolates Resistant to Nalidixic Acid
|
Mean of MIC50
|
Antibiotics
|
Concentrations of Antibiotics and Number of Isolates Resistant to Each Antibiotic at Each Concentration
|
|
0.125
|
0.25
|
0.5
|
1
|
2
|
4
|
8
|
16
|
| 11.56 |
Cefprozil |
0 |
0 |
0 |
0 |
0 |
2 |
2 |
5 |
| 2.78 |
Ceftazidime |
0 |
0 |
0 |
1 |
4 |
4 |
0 |
0 |
| 1.45 |
Chloramphenicol |
0 |
0 |
0 |
5 |
4 |
0 |
0 |
0 |
| 0.64 |
Gentamicin |
0 |
3 |
2 |
4 |
0 |
0 |
0 |
0 |
| 9.33 |
Minocycline |
0 |
0 |
0 |
0 |
0 |
3 |
3 |
3 |
| 1.22 |
Lomefloxacin |
0 |
0 |
2 |
4 |
3 |
0 |
0 |
0 |
| 1.28 |
Levofloxacin |
0 |
0 |
5 |
1 |
2 |
1 |
0 |
0 |
| 3.67 |
Ciprofloxacin |
0 |
0 |
0 |
1 |
4 |
2 |
2 |
0 |
| 2.67 |
Sparfloxacin |
0 |
0 |
0 |
2 |
5 |
1 |
1 |
0 |
| 5.78 |
Ofloxacin |
0 |
0 |
0 |
2 |
1 |
2 |
3 |
1 |
Note. MIC: Minimum inhibitory concentration.
Minimum Inhibitory Concentrations Against S. paratyphi Isolates Susceptible to Nalidixic Acid
Based on the obtained data (Table 7), chloramphenicol (MIC50mean = 0.19 μg/mL), gentamicin (MIC50mean = 0.48 μg/mL), and ceftazidime (MIC50mean = 0.63 μg/mL) were the most effective antibiotics against 12 S. paratyphi isolates sensitive to nalidixic acid, and all isolates were sensitive to these antibiotics. On the other hand, all isolates were resistant to cefprozil and minocycline. Moreover, most fluoroquinolones represented high effectiveness against these isolates, and the MIC50mean was 0.51 μg/mL, 0.54 μg/mL, 0.44 μg/mL, and 0.29 μg/mL for lomefloxacin, levofloxacin, ciprofloxacin, and ofloxacin, respectively.
Table 7.
MIC50 in Salmonella paratyphi Isolates Susceptible to Nalidixic Acid
|
Mean of MIC50
|
Antibiotics
|
Concentrations of Antibiotics and Number of Isolates Susceptible to Each Antibiotic at Each Concentration
|
|
0.125
|
0.25
|
0.5
|
1
|
2
|
4
|
8
|
16
|
| 14 |
Cefprozil |
0 |
0 |
0 |
0 |
0 |
0 |
3 |
9 |
| 0.63 |
Ceftazidime |
0 |
4 |
3 |
5 |
0 |
0 |
0 |
0 |
| 0.19 |
Chloramphenicol |
6 |
6 |
0 |
0 |
0 |
0 |
0 |
0 |
| 0.48 |
Gentamicin |
0 |
3 |
8 |
1 |
0 |
0 |
0 |
0 |
| 9.5 |
Minocycline |
0 |
0 |
0 |
0 |
1 |
2 |
5 |
4 |
| 0.51 |
Lomefloxacin |
1 |
4 |
4 |
3 |
0 |
0 |
0 |
0 |
| 0.54 |
Levofloxacin |
0 |
4 |
5 |
3 |
0 |
0 |
0 |
0 |
| 0.44 |
Ciprofloxacin |
2 |
6 |
1 |
3 |
0 |
0 |
0 |
0 |
| 1.07 |
Sparfloxacin |
1 |
1 |
3 |
3 |
4 |
0 |
0 |
0 |
| 0.29 |
Ofloxacin |
4 |
4 |
4 |
0 |
0 |
0 |
0 |
0 |
Note. MIC: Minimum inhibitory concentration.
Minimum Inhibitory Concentrations Against Salmonella paratyphi Isolates Resistant to Nalidixic Acid
The results (Table 8) showed that gentamicin (MIC50mean = 0.75 μg/mL) was the most effective antibiotic against 11 S. paratyphi isolates resistant to nalidixic acid, and all isolates were sensitive to this antibiotic. Among the quinolones, levofloxacin and lomefloxacin (MIC50mean = 1.14 and MIC50mean = 1.27 μg/mL, respectively) demonstrated moderate efficacy against these isolates. However, 9 (82%) and 8 (73%) isolates were sensitive to levofloxacin and lomefloxacin, respectively. On the other hand, 4 isolates were sensitive to chloramphenicol and sparfloxacin (36%), and three isolates were sensitive to ofloxacin (27%). Finally, all isolates were resistant to cefprozil, ceftazidime, minocycline, and ciprofloxacin.
Table 8.
MIC50 in Salmonella paratyphi Isolates Resistant to Nalidixic Acid
|
Mean of MIC50
|
Antibiotics
|
Concentrations of Antibiotics and Number of Isolates Resistant to Each Antibiotic at Each Concentration
|
|
0.125
|
0.25
|
0.5
|
1
|
2
|
4
|
8
|
16
|
| 15.27 |
Cefprozil |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
10 |
| 8.73 |
Ceftazidime |
0 |
0 |
0 |
0 |
0 |
2 |
7 |
2 |
| 2 |
Chloramphenicol |
0 |
0 |
0 |
4 |
5 |
2 |
0 |
0 |
| 0.75 |
Gentamicin |
0 |
1 |
4 |
6 |
0 |
0 |
0 |
0 |
| 10.18 |
Minocycline |
0 |
0 |
0 |
0 |
0 |
4 |
2 |
5 |
| 1.27 |
Lomefloxacin |
0 |
0 |
0 |
8 |
3 |
0 |
0 |
0 |
| 1.14 |
Levofloxacin |
0 |
0 |
5 |
4 |
1 |
1 |
0 |
0 |
| 6.73 |
Ciprofloxacin |
0 |
0 |
0 |
0 |
3 |
3 |
3 |
2 |
| 2.36 |
Sparfloxacin |
0 |
0 |
0 |
4 |
5 |
1 |
1 |
0 |
| 5.18 |
Ofloxacin |
0 |
0 |
0 |
3 |
3 |
2 |
1 |
2 |
Note. MIC: Minimum inhibitory concentration.
Discussion
Typhoid fever continues to pose a real threat to human health in developing countries (17,24-26). Although its severity varies between regions, this disease causes approximately 21.6 million cases of infection and 216.55 deaths worldwide annually (17).
Unacceptably high rates of any infectious disease or a significant increase in the number of infections with this disease strongly motivate greater efforts to prevent it, whether by following health guidelines or performing vaccination campaigns.
The resistance of S. typhi to chloramphenicol, amoxicillin, and co-trimoxazole is a challenge to applied therapeutic regimens. Fluoroquinolones have emerged as experimental therapeutic drugs for this disease. Typically, all S. typhi isolates are susceptible to ciprofloxacin when using the disk diffusion method approved by the International Council for Laboratory Standards (27). The excessive use of fluoroquinolones in the treatment of typhoid fever has led to an increase in the dose of ciprofloxacin to be used in treatment, demonstrating an increase in the MIC of this drug. Reports show a lack of susceptibility of S. typhi isolates to ciprofloxacin in Great Britain, as well as India and its neighboring countries (28,29). Low resistance to ciprofloxacin may result in delayed patient response to treatment or incomplete recovery and serious consequences for successful treatment. Studies suggest the possibility of considering the presence of resistance to nalidixic acid in the disk diffusion method as an indirect indicator of the presence of resistance to quinolones (30). Many studies have shown resistance to several antibiotics used as first-line treatments for S. typhi (31,32).
In recent decades, Salmonella, especially S. typhi, has been able to rapidly develop resistance to antibiotics such as ampicillin, chloramphenicol, and cotrimoxazole, and even to ciprofloxacin (32). MDR enteric fever remains a major problem in all countries (33,34).
In 1993, a significant increase was observed in the number of MDR strains of Salmonella in developing countries, in addition to their dramatic resistance to nalidixic acid. In 1998, after 5 years of uncontrolled use of ofloxacin and ciprofloxacin, the proportion of drug-resistant isolates increased, with 87% of strains being resistant to nalidixic acid; this percentage increased to 97% in 2004 (35). The combination of MDR and nalidixic acid resistance is a major problem in such countries, as this reduces treatment options for typhoid fever patients. The treatment response in patients infected with nalidixic acid-resistant strains is poor, treatment failure rates are high (up to 36%), and the fecal burden of these strains is prolonged for long periods when treated with older generations of quinolones such as ofloxacin (36). Reports from Nepal, India, and Bangladesh of a significant increase in the number of ciprofloxacin-resistant strains sparked the worst drug resistance problem in Asia (37-39). The emergence of isolates of S. typhi that represent resistance to ciprofloxacin and third-generation cephalosporin drugs is of great interest to all clinicians in developing countries (39-41).
Although aminoglycosides are not recommended for the treatment of typhoid fever, this group has shown significant activity against Salmonella isolates in vitro. In addition, a number of cases of gentamicin response have been reported in patients resistant to ciprofloxacin (41).
However, our findings revealed that the most effective drugs against Salmonella isolates isolated from meat, chicken eggs, or milk were gentamicin (an aminoglycoside) and chloramphenicol and ceftazidime (third-generation cephalosporins), whether these isolates are sensitive or resistant to nalidixic acid. However, neither cefprozil (a first-generation cephalosporin) nor minocycline demonstrated any significant activity against all isolates. These results are in line with those of Mandal et al (42).
As for fluoroquinolones, our results confirmed that the best drugs in this drug group were levofloxacin (a third-generation fluoroquinolone) and lomefloxacin (a second-generation fluoroquinolone), and to a lesser extent ofloxacin. Conversely, sparfloxacin (the third generation) was the least effective fluoroquinolone. Ciprofloxacin (second generation) showed good activity against isolates sensitive to nalidixic acid only. The results related to this group conform to those that have been published in this regard (43).
Conclusion
MDR was not revealed in this study, in vitro. However, varying activities of drugs within the same drug group were observed. Although gentamicin was the most effective drug, it cannot be applied to clinical therapy in any way since aminoglycosides are drugs that specifically affect extracellular bacteria. In addition, despite the differences in fluoroquinolone effects, they generally show good effectiveness. Moreover, third-generation cephalosporins still have significant efficacy against Salmonella in vitro.
Authors’ Contribution
Conceptualization: Ayman Al-Mariri and Mazen Safi.
Data curation: Mazen Safi and Ayman Al-Mariri.
Formal analysis: Mazen Safi and Bassam Al Balaa.
Investigation: Mazen Safi, Samah Qasem, and Laila Al Hallab.
Methodology: Mazen Safi and Ayman Al-Mariri.
Project administration: Mazen Safi, Bassam Al Balaa, and Ayman Al-Mariri.
Resources: Mazen Safi, Bassam Al Balaa, and Ayman Al-Mariri.
Supervision: Mazen Safi and Ayman Al-Mariri.
Validation: Mazen Safi and Ayman Al-Mariri.
Visualization: Mazen Safi, Samah Qasem, and Laila Al Hallab.
Writing-original draft: Mazen Safi and Bassam Al Balaa.
Writing-review & editing: Mazen Safi.
Competing Interests
The authors declare that there are no competing interests.
Ethical Approval
This study was conducted in vitro. There is no experiment conducted on humans or live animals.
Funding
None.
References
- Kim C, Goucher GR, Tadesse BT, Lee W, Abbas K, Kim JH. Associations of water, sanitation, and hygiene with typhoid fever in case-control studies: a systematic review and meta-analysis. BMC Infect Dis 2023; 23(1):562. doi: 10.1186/s12879-023-08452-0 [Crossref] [ Google Scholar]
- Kirchhelle C, Dyson ZA, Dougan G. A biohistorical perspective of typhoid and antimicrobial resistance. Clin Infect Dis 2019; 69(Suppl 5):S388-94. doi: 10.1093/cid/ciz556 [Crossref] [ Google Scholar]
- Besser JM. Salmonella epidemiology: a whirlwind of change. Food Microbiol 2018; 71:55-9. doi: 10.1016/j.fm.2017.08.018 [Crossref] [ Google Scholar]
- Fangtham M, Wilde H. Emergence of Salmonella paratyphi A as a major cause of enteric fever: need for early detection, preventive measures, and effective vaccines. J Travel Med 2008; 15(5):344-50. doi: 10.1111/j.1708-8305.2008.00237.x [Crossref] [ Google Scholar]
- Gedeno K, Hailegebreal G, Tanga BM, Sulayeman M, Sori T. Epidemiological investigations of Salmonella and Escherichia coli associated morbidity and mortality in layer chickens in Hawassa city, Southern Ethiopia. Heliyon 2022; 8(12):e12302. doi: 10.1016/j.heliyon.2022.e12302 [Crossref] [ Google Scholar]
- Holschbach CL, Peek SF. Salmonella in dairy cattle. Vet Clin North Am Food Anim Pract 2018; 34(1):133-54. doi: 10.1016/j.cvfa.2017.10.005 [Crossref] [ Google Scholar]
- Glenn LM, Lindsey RL, Folster JP, Pecic G, Boerlin P, Gilmour MW. Antimicrobial resistance genes in multidrug-resistant Salmonella enterica isolated from animals, retail meats, and humans in the United States and Canada. Microb Drug Resist 2013; 19(3):175-84. doi: 10.1089/mdr.2012.0177 [Crossref] [ Google Scholar]
- Wang M, Qazi IH, Wang L, Zhou G, Han H. Salmonella virulence and immune escape. Microorganisms 2020; 8(3):407. doi: 10.3390/microorganisms8030407 [Crossref] [ Google Scholar]
- Ryan KJ, Ray CG, Sherris JC. Sherris Medical Microbiology. 8th ed. McGraw-Hill; 2022.
- Carvajal A, Kramer M, Argüello H. Salmonella control in swine: a thoughtful discussion of the pre- and post-harvest control approaches in industrialized countries. Animals (Basel) 2024; 14(7):1035. doi: 10.3390/ani14071035 [Crossref] [ Google Scholar]
- Popoff MY, Bockemühl J, Brenner FW. Supplement 1998 (no 42) to the Kauffmann-White scheme. Res Microbiol 2000; 151(1):63-5. doi: 10.1016/s0923-2508(00)00126-1 [Crossref] [ Google Scholar]
- Galen JE, Wahid R, Buskirk AD. Strategies for enhancement of live-attenuated Salmonella-based carrier vaccine immunogenicity. Vaccines (Basel) 2021; 9(2):162. doi: 10.3390/vaccines9020162 [Crossref] [ Google Scholar]
- Woodward TE, Smadel JE, Ley HL Jr, Green R, Mankikar DS. Preliminary report on the beneficial effect of chloromycetin in the treatment of typhoid fever. Ann Intern Med 1948; 29(1):131-4. doi: 10.7326/0003-4819-29-1-131 [Crossref] [ Google Scholar]
- Olarte J, Galindo E. Salmonella typhi resistant to chloramphenicol, ampicillin, and other antimicrobial agents: strains isolated during an extensive typhoid fever epidemic in Mexico. Antimicrob Agents Chemother 1973; 4(6):597-601. doi: 10.1128/aac.4.6.597 [Crossref] [ Google Scholar]
- Biswas M, Biswas S, Gupta B, Mascellino MT, Rakshit A, Chakraborty B. Changing paradigms in antibiotic resistance in Salmonella species with focus on fluoroquinolone resistance: a 5-year retrospective study of enteric fever in a tertiary care hospital in Kolkata, India. Antibiotics (Basel) 2022; 11(10):1308. doi: 10.3390/antibiotics11101308 [Crossref] [ Google Scholar]
- Wain J, Kidgell C. The emergence of multidrug resistance to antimicrobial agents for the treatment of typhoid fever. Trans R Soc Trop Med Hyg 2004; 98(7):423-30. doi: 10.1016/j.trstmh.2003.10.015 [Crossref] [ Google Scholar]
- Bhan MK, Bahl R, Bhatnagar S. Typhoid and paratyphoid fever. Lancet 2005; 366(9487):749-62. doi: 10.1016/s0140-6736(05)67181-4 [Crossref] [ Google Scholar]
- Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med 2002; 347(22):1770-82. doi: 10.1056/NEJMra020201 [Crossref] [ Google Scholar]
- Murray PR. Manual of Clinical Microbiology. 8th ed. Washington, DC: ASM Press; 2003. p. 2310.
- Crump JA, Ram PK, Gupta SK, Miller MA, Mintz ED. Part I Analysis of data gaps pertaining to Salmonella enterica serotype Typhi infections in low and medium human development index countries, 1984-2005. Epidemiol Infect 2008; 136(4):436-48. doi: 10.1017/s0950268807009338 [Crossref] [ Google Scholar]
- Safi M, Al Balaa B, Qasem S, Al-Hallab L, Al-Mariri A. Isolation and Identification of Salmonella typhimurium in retail meat sources by multiplex polymerase chain reaction. Avicenna J Clin Microbiol Infect 2022; 9(4):152-6. doi: 10.34172/ajcmi.2022.3402 [Crossref] [ Google Scholar]
- Safi M, Al-Hallab L, Al-Abras R, Khawajkiah M, Kherbik H, Al-Mariri A. Efficacy of some antibiotics and essential oils against Acinetobacter baumannii: an in vitro study. Avicenna J Clin Microbiol Infect 2020; 7(1):1-7. doi: 10.34172/ajcmi.2020.01 [Crossref] [ Google Scholar]
- National Committee for Clinical Laboratory Standards (NCCLS). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. 6th ed. Approved Standard M7-A6. Wayne, PA: NCCLS; 2003.
- Batool R, Qamar ZH, Salam RA, Yousafzai MT, Ashorn P, Qamar FN. Efficacy of typhoid vaccines against culture-confirmed Salmonella Typhi in typhoid endemic countries: a systematic review and meta-analysis. Lancet Glob Health 2024; 12(4):e589-98. doi: 10.1016/s2214-109x(23)00606-x [Crossref] [ Google Scholar]
- Mweu E, English M. Typhoid fever in children in Africa. Trop Med Int Health 2008; 13(4):532-40. doi: 10.1111/j.1365-3156.2008.02031.x [Crossref] [ Google Scholar]
- Nga TV, Duy PT, Lan NP, Chau NV, Baker S. The control of typhoid fever in Vietnam. Am J Trop Med Hyg 2018; 99(3 Suppl):72-8. doi: 10.4269/ajtmh.18-0035 [Crossref] [ Google Scholar]
- National Committee for Clinical Laboratory Standards (NCCLS). Performance Standards for Antimicrobial Disc Susceptibility Tests. 7th ed. Approved Standards M2-A6. Wayne, PA: NCCLS; 2000.
- Dimitrov T, Dashti AA, Albaksami O, Udo EE, Jadaon MM, Albert MJ. Ciprofloxacin-resistant Salmonella enterica serovar Typhi from Kuwait with novel mutations in gyrA and parC genes. J Clin Microbiol 2009; 47(1):208-11. doi: 10.1128/jcm.01161-08 [Crossref] [ Google Scholar]
- Ikram S, Hussain S, Imtiaz A, Khan MD. Nalidixic acid as a predictor of ciprofloxacin susceptibility in typhoid fever. J Islamabad Med Dent Coll 2015; 4(3):118-21. [ Google Scholar]
- Ray P, Sharma J, Marak RS, Garg RK. Predictive efficacy of nalidixic acid resistance as a marker of fluoroquinolone resistance in Salmonella enterica var Typhi. Indian J Med Res 2006; 124(1):105-8. [ Google Scholar]
- Arjyal A, Basnyat B, Koirala S, Karkey A, Dongol S, Agrawaal KK. Gatifloxacin versus chloramphenicol for uncomplicated enteric fever: an open-label, randomised, controlled trial. Lancet Infect Dis 2011; 11(6):445-54. doi: 10.1016/s1473-3099(11)70089-5 [Crossref] [ Google Scholar]
- Mandal S, DebMandal M, Pal NK, Saha K. Inhibitory and killing activities of black tea (Camellia sinensis) extract against Salmonella enterica serovar Typhi and Vibrio cholerae O1 biotype El Tor serotype Ogawa isolates. Jundishapur J Microbiol 2011; 4(2):115-21. [ Google Scholar]
- Kalra SP, Naithani N, Mehta SR, Swamy AJ. Current trends in the management of typhoid fever. Med J Armed Forces India 2003; 59(2):130-5. doi: 10.1016/s0377-1237(03)80060-6 [Crossref] [ Google Scholar]
- Threlfall EJ, Day M, de Pinna E, Lewis H, Lawrence J. Drug-resistant enteric fever in the UK. Lancet 2006; 367(9522):1576. doi: 10.1016/s0140-6736(06)68691-1 [Crossref] [ Google Scholar]
- Chau TT, Campbell JI, Galindo CM, Van Minh Hoang N, Diep TS, Nga TT. Antimicrobial drug resistance of Salmonella enterica serovar Typhi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicrob Agents Chemother 2007; 51(12):4315-23. doi: 10.1128/aac.00294-07 [Crossref] [ Google Scholar]
- Parry CM, Ho VA, Thi Phuong L, Bay PV, Lanh MN, Thanh Tung L. Randomized controlled comparison of ofloxacin, azithromycin, and an ofloxacin-azithromycin combination for treatment of multidrug-resistant and nalidixic acid-resistant typhoid fever. Antimicrob Agents Chemother 2007; 51(3):819-25. doi: 10.1128/aac.00447-06 [Crossref] [ Google Scholar]
- Renuka K, Sood S, Das BK, Kapil A. High-level ciprofloxacin resistance in Salmonella enterica serotype Typhi in India. J Med Microbiol 2005; 54(Pt 10):999-1000. doi: 10.1099/jmm.0.45966-0 [Crossref] [ Google Scholar]
- Gaind R, Paglietti B, Murgia M, Dawar R, Uzzau S, Cappuccinelli P. Molecular characterization of ciprofloxacin-resistant Salmonella enterica serovar Typhi and Paratyphi A causing enteric fever in India. J Antimicrob Chemother 2006; 58(6):1139-44. doi: 10.1093/jac/dkl391 [Crossref] [ Google Scholar]
- Saha SK, Darmstadt GL, Baqui AH, Crook DW, Islam MN, Islam M. Molecular basis of resistance displayed by highly ciprofloxacin-resistant Salmonella enterica serovar Typhi in Bangladesh. J Clin Microbiol 2006; 44(10):3811-3. doi: 10.1128/jcm.01197-06 [Crossref] [ Google Scholar]
- Chowdhury SR, Ahamed Z, Roy K, Al Noman A, Haroon RM, Mondol KC. Emerging threats of antibiotic resistance in Salmonella Typhi and Salmonella paratyphi A among enteric fever cases of Dhaka, Bangladesh. Afr J Bacteriol Res 2022; 14(1):8-15. doi: 10.5897/jbr2021.0340 [Crossref] [ Google Scholar]
- Prabha Adhikari MR, Baliga S. Ciprofloxacin-resistant typhoid with incomplete response to cefotaxime. J Assoc Physicians India 2002; 50:428-9. [ Google Scholar]
- Mandal S, Mandal MD, Pal NK. In vitro activity of gentamicin and amikacin against Salmonella enterica serovar Typhi: a search for a treatment regimen for typhoid fever. East Mediterr Health J 2009; 15(2):264-8. [ Google Scholar]
- Joshi S, Amarnath SK. Fluoroquinolone resistance in Salmonella typhi and S paratyphi A in Bangalore, India. Trans R Soc Trop Med Hyg 2007; 101(3):308-10. doi: 10.1016/j.trstmh.2006.05.009 [Crossref] [ Google Scholar]