Avicenna Journal of Clinical Microbiology and Infection. 11(3):100-106.
doi: 10.34172/ajcmi.3557
Original Article
Implications of Biofilm-Producing Organisms Among Bacteria Isolated From Ear-, Nose-, and Throat-Infected Patients
Oluwabusayomi Roseline Ademakinwa 1  , Adekunle Adeyemo 2, 3
, Adekunle Adeyemo 2, 3  , Samuel Oluyomi Ayodele 3, *
, Samuel Oluyomi Ayodele 3, *  , Akumbu Sylvia Nwamuo 3, Anthonia Olufunke Oluduro 1
, Akumbu Sylvia Nwamuo 3, Anthonia Olufunke Oluduro 1
Author information:
1Department of Microbiology, Faculty of Sciences, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria
2Department of Otorhinolaryngology, Faculty of Clinical Sciences, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria
3Department of Otorhinolaryngology-Head and Neck Surgery, Obafemi Awolowo University Teaching Hospital Complex, IleIfe, Osun State, Nigeria
Abstract
Background: Bacteria resistant to antimicrobial agents have remained a major challenge in public health, and bacterial-producing biofilm is one of the main causes of antibiotic resistance, especially in upper respiratory tract infections (URTI). This study aimed at determining the antibiotic resistance pattern and formation of biofilms in bacteria causing ear, nose, and throat (ENT) infections in our study population.
Methods: One hundred and fifty samples, including ear (n=87), nasal discharge (22), throat swab (8), and surgical sample (33) (aspirate and tissue), were screened and analyzed using the culture technique, direct microscopy, and bacteria identification with an API 20E strip. The antibiotic susceptibility testing of the isolates was performed with Kirby-Bauer’s disk diffusion techniques and interpreted based on the Clinical and Laboratory Standard Institute guidelines. The biofilm-producing organisms (BPOs) were determined by using the tube method technique.
Results: A total of 192 isolates were recovered (60% gram-positive and 40% gram-negative bacteria). Eighty-three (43.2%) of recovered isolates were multidrug-resistant (MDR) to antibiotics tested, and 60 (75%) isolates from MDR isolates were BPOs.
Conclusion: Biofilm-producing bacteria have higher tendencies to dominate in body-infected tissues other than the discharges being produced; therefore, tissue biopsy for culture and sensitivity should be considered more appropriate where visible, especially when confronted with hard-to-treat infections in ENT clinical settings.
Keywords: Ear, nose and throat infection, Antibiotic resistance, Biofilm-producing organism
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Ademakinwa OR, Adeyemo A, Ayodele SO, Nwamuo AS, Oluduro AO. Implications of biofilm-producing organisms among bacteria isolated from ear-, nose-, and throat-infected patients. Avicenna J Clin Microbiol Infect. 2024; 11(3):100-106. doi:10.34172/ajcmi.3557
Introduction
Antimicrobial resistance (AMR) is a significant public health threat, as admitted by the World Health Organization (WHO) (1). The extent of this AMR and the selection of multidrug-resistant (MDR) pathogens have been reported to be a result of non-compliance with proper infection control methods, unfounded use of antibiotics, availability of antibiotics without a prescription, and counterfeit products of dubious quality (2). Upper respiratory tract infections (URTI) are commonly treated with antibiotics based on the known sensitivity patterns of the common causative pathogens encountered in specific regions (3). Antimicrobial agents remain the backbone of infectious disease treatment; however, the unjustifiable uses of antibiotics in many countries have evolved into the emergence of MDR microorganisms (4). Infections become chronic, incalcitrant, or present with complications when treated with antibiotics that are not responsive to infective agents. The abuse of antimicrobial agents is well known to create pressure on the selection of appropriate drugs and resultantly increase the capability of microbes to restrain from being attacked (5). Antibiotic resistance leads to higher medical expenses, prolonged hospital stays, and increased mortality rates (2). Studies have reported an alarming increase in the occurrence of antibiotic-resistant bacteria that include beta-lactam-resistant strains of common pathogens as well as macrolides and fluoroquinolone-resistant strains isolated from URTIs (6,7).
An important element that contributes to AMR is biofilm production by bacteria. The formation of biofilm has been reported as one of the factors causing antibiotic resistance, particularly in URTIs. Bacterial biofilms are naturally resistant to antibioticsdue to the fact that some antibiotics are unable to reach the depths of the biofilm; some cells in biofilms grow slowly or not at all, likely due to nutrient limitation, and certain cells in the biofilms may adopt a unique and safeguarded biofilm phenotype (8).
Studies have identified bacterial biofilm as the main cause of antibiotic resistance in URTIs (9). Biofilm can form on moist biotic and abiotic surfaces, making them common for infection of the ear, nose, and throat (ENT). Bacterial biofilms are known to be “influencers of infections”, especially in patients suffering from rhinosinusitis and otitis media as a result of their propensity to form biofilms in sinuses and adenoid tissues (9–13). Most bacteria (more than 99%) produce biofilms, which can lead to dangerous, incurable illnesses. The bacteria in biofilms interact with each other through molecular mechanisms that enable some cells to resist antibiotics and host immune defenses, hence increasing the likelihood that ENT infections would recur or persist (13).
It has been estimated that at least 25% of cases of chronic rhinosinusitis are caused by biofilm formation. (14) Many microorganisms such as Streptococcus spp., Staphylococcus aureus, Corynebacterium argentoratense, and Micrococcus luteus present in the respiratory tract have been reported to easily produce biofilms. S. aureus strains have been identified intracellularly and in the sub-mucosa of adult patients with chronic rhinosinusitis undergoing endoscopic sinus surgery (15). Microorganisms such as S. aureus,Haemophilus influenza, Pseudomonas aeruginosa,and fungus have been isolated and identified as bacterial biofilms from chronic rhinosinusitis (9).
Despite the knowledge of bacteria as the major etiological agents of URTI and the roles of antibiotic resistance and formation of biofilms in the pathogenesis of URTI, local studies have not focused on biofilm formation potentials in bacteria causing ENT infections in West Africa. Therefore, this study seeks to establish the antibiotic sensitivity pattern and formation of biofilms in bacteria causing ENT infections in our study population and to determine the contribution of biofilm formation to MDR agents and the most effective choice of empiric antibiotics.
Materials and Methods
Sampling Population/Ethical Clearance
Participants with a clinical diagnosis of ENT infections attending the Otorhinolaryngology (ENT) Clinic in Obafemi Awolowo University Teaching Hospitals Complex (OAUTHC), Ile-Ife, Nigeria during the study period formed the study population. Ethical clearance (with protocol number ERC/2018/06/14) was duly obtained from the Ethical and Research Committee of the OAUTHC. In addition, informed consent was obtained from the patient or guardian of the patient as appropriate.
Sample Collection
ENT samples in the form of discharges, aspirates, and infected tissues were collected at the Otorhinolaryngology Clinic and Main Theatre of OAUTHC. Each sample was collected aseptically using sterile swab sticks or sterile bottles as appropriate by a medical doctor who has been pre-trained on the study protocol in the otorhinolaryngology clinic and operating theatre. The samples were collected into freshly prepared sterile transport media (thioglycolate media), properly labelled (with study number, date, gender, age, and time), and immediately transported for bacteriological analysis at the Microbiology Department of Obafemi Awolowo University, Ile-Ife, Nigeria.
Bacteria Isolation and Identification
The sample collected in the transport media was incubated over 24 hours at 37 ºC. The incubated culture was then inoculated separately on sterile blood agar, nutrient agar, MacConkey agar, and Mannitol salt agar (Lab M Ltd., UK) by the streak plate method for discrete colonies and incubated at 37 °C for 24 hours. The organisms were purified by successive subculturing on a nutrient agar plate. The isolates were identified by morphological and physiological characteristics according to Bergey’s Manual of Determinative (16). Furthermore, the isolates were identified by using the MICROBACTTM identification kits 24E (Oxoid Ltd., UK) for Gram-negative, and the STPY gene was used to identify S. aureus using the polymerase chain reaction method.
Antibiotic Susceptibility Test and Biofilm Formation
The antibiotic susceptibility profile of the isolates was determined on Mueller-Hinton agar (MHA; Lab M Ltd., UK) according to Kirby-Bauer’s disc diffusion technique (17). The antibiotic discs, including single (Oxoid Ltd., Basingstoke, Hampshire, England) and combined (Abtek Biological Ltd., UK) discs of varying and specific concentrations, were employed and aseptically placed on the inoculated MHA plate with sterile forceps. The antibiotic discs were properly placed on MHA plates, seeded with standardized (106 CFU/mL of 0.5 McFarland Standard) inoculum, and the plates were incubated at 37 °C for 18–24 hours, after which the diameter of zones of inhibition was compared with the Clinical and Laboratory Standard Institute (18) chart of interpretative zones as sensitive, resistance, and intermediate resistance. The isolates were described as resistant to multiple antibiotics when they were resistant to ≥ 3 separate classes of the tested antibiotics. The qualitative method for the biofilm formation of the isolates was performed using the tube method. A loop full of test organisms was inoculated in 10 mL of nutrient broth (Lab M Ltd., UK) with 1% glucose in test tubes. After incubation, the tubes were decanted and washed with the use of the phosphate buffer saline (pH = 7.3). The tubes were dried and then stained with crystal violet (0.1%). Deionized water was used to wash off excess stains, and the tubes were dried in an inverted position. The tube method was scored in line with the results from the control strains. As depicted in Figure 1, biofilm formation was considered positive when a visible film lined the wall and the bottom of the tube. No biofilm was produced when the tube was clear, implying that the wall and bottom of the tube were not lined by any visible film (19).

Figure 1.
An Image of Biofilm-Producing Bacteria Recovered From ENT Infections. Note. ENT: Ear, nose, and throat
.
An Image of Biofilm-Producing Bacteria Recovered From ENT Infections. Note. ENT: Ear, nose, and throat
Statistical Analysis
The data collected for analysis included patient sociodemographic details, previous exposure to antibiotics in current ENT disease, nature of the specimen, nature of isolates, antibiotic resistance, and biofilm production. Statistical Product and Service Solutions Statistics (version 22) was utilized to perform statistical analysis with the level of statistical significance set at P ≤ 0.05.
Results
One hundred and fifty samples, including ear (n = 87), nasal (n = 22) throat (n = 8), and ENT surgical aspirates and tissues (n = 33), were obtained from 150 patients [77 (51.3%) females and 73 (48.7%) males] diagnosed with various ENT infections. The age range of 0–5 years had the highest population in this study, followed by age ≥ 46, while 41–45 years had the lowest population. Table 1 provides a summary of the collected samples.
Table 1.
Nature of Samples Collected From Their Site of Infection
|
Nature of the Sample
|
Site of Infection
|
Ear infection
n=88 (58.7%)
|
Nose Infection
n=46 (30.6%)
|
Throat infection
n=16 (30.6%)
|
Total
n=150 (100)
|
| Discharge |
87 (98.9) |
22 (47.8) |
8 (50.0) |
117 (78) |
| Tissue |
1 (1.1) |
24 (52.2) |
8 (50.0) |
33 (22) |
As outlined in Tables 2 and 3, a total of 15 different bacterial species were identified from 192 bacteria isolated from 150 samples [110 (73.3%) mono-bacterial were cultured from collected samples, and poly-bacterial culture was found in 40 (26.70%) samples]. These species were Proteus spp., P. aeruginosa, Klebsiella pneumoniae, Escherichia coli, Citrobacter freundii, Enterobacter cloacae, Serratia spp., Salmonella spp., Acinetobacter baumannii, Staphylococcus spp., Bacillus spp., Corynebacterium spp., Streptococcus spp., Micrococcus spp., and Enterococcus spp. Antibiotic susceptibility showed meropenem, vancomycin, and ofloxacin as the most effective antibiotics, while the isolates are more resistant to commonly used empirical antibiotics such as augmentin, cotrimoxazole, ceftazidime, cefuroxime, gentamycin, erythromycin, chloramphenicol, and ampicillin.
Table 2.
Antibiotic Susceptibility Pattern of Gram-positive Bacteria Cultured From Ear, Nose, and Throat Infection Samples
|
Antibiotic (µg)
|
No. of Isolates
|
Number of Isolate Occurrence (%)
|
|
Susceptibility
|
Intermediate
|
Resistance
|
| VAN |
115 |
115 (100) |
0 (0) |
0 (0) |
| MEM |
115 |
114 (99) |
1 (1) |
0 (0) |
| OFL |
115 |
108 (94) |
6 (5) |
1 (1) |
| AUG |
115 |
62 (54) |
0 (0) |
53 (46) |
| AMP |
115 |
62 (54) |
2 (1.7) |
51 (44) |
| CRX |
115 |
53 (46) |
11 (10) |
51 (44) |
| COT |
115 |
65 (56.5) |
2 (1.7) |
48 (41.7) |
| GEN |
115 |
64 (56) |
22 (19) |
29 (25) |
| CAZ |
115 |
42 (36.5) |
16 (13.9) |
57 (49.6) |
| TET |
115 |
49 (42.6) |
26 (22.6) |
40 (34.8) |
| ERY |
115 |
46 (40) |
42 (36.5) |
27 (23.5) |
Note. CAZ:Ceftazidime (30 µg); CRX: Cefuroxime (30 µg); OFL: Ofloxacin (5 µg); AUG: Augmentin (30 µg); NIT: Nitrofurantoin (300 µg); CPR: Ciprofloxacin (5 µg); GEN: Gentamycin (10 µg); CXM: Cefixime (5 µg); CHL: Chloramphenicol (30 µg); TET: Tetracycline (30 µg); MEM: Meropenem (10 µg); FOX: Cefoxitin (30 µg); COT: Cotrimoxazole (25 µg); CTX: Cefotaxime (30 µg); CTR: Ceftriaxone (30 µg); AMK: Amikacin (30 µg); ERY: Erythromycin; VAN: Vancomycin; AMP: Ampicillin (10 µg).
Table 3.
Antibiotic Susceptibility Pattern of Gram-negative Bacterial Cultured From Ear, Nose, and Throat Infection Samples
|
Antibiotics (µg)
|
No. of Isolates
|
Number of Isolate Occurrence (%)
|
|
Susceptibility
|
Intermediate
|
Resistance
|
| MEM |
77 |
77 (100) |
0 (0) |
0 (0) |
| OFL |
77 |
69 (89.6) |
1 (1.3) |
7 (9.1) |
| CXM |
77 |
49 (63.6) |
8 (10.4) |
20 (26) |
| NIT |
77 |
48 (62.3) |
11 (14.3) |
18 (23.4) |
| CPR |
77 |
64 (83.1) |
6 (7.8) |
7 (9.1) |
| FOX |
77 |
46 (59.7) |
11 (14.3) |
20 (26) |
| GEN |
77 |
48 (62.3) |
9 (11.7) |
20 (26) |
| CHL |
77 |
41 (53.2) |
14 (18.2) |
22 (28.6) |
| CTR |
77 |
38 (49.4) |
25 (32.5) |
14 (18.2) |
| AMK |
77 |
33 (42.9) |
11 (14.3) |
23 (29.9) |
| COT |
77 |
33 (42.9) |
8 (10.4) |
36 (46.8) |
| CTX |
77 |
28 (36.4) |
30 (39) |
19 (24.7) |
| CAZ |
77 |
26 (33.8) |
26 (33.8) |
25 (32.5) |
| CRX |
77 |
23 (29.9) |
19 (24.7) |
35 (45.5) |
| AUG |
77 |
22 (28.6) |
26 (33.8) |
29 (37.7) |
| TET |
77 |
26 (33.8) |
9 (11.7) |
42 (54.5) |
Based on the results (Table 4), 192 bacteria were recovered from collected samples, 113 isolates were isolated from ear infections, and 51 (45.1%) were multiple antibiotic resistant (MAR). In addition, 46 isolates were recovered from nose infection, and 20 (43.5%) were MAR. Twenty-three isolates were recovered from the throat, while 12 (52.2%) were MAR.
Table 4.
Frequency of Multidrug Gram-negative and Gram-Positive Isolates Cultured ENT Infection Samples
|
Site of infection
|
Ear Infection
|
Nose Infection
|
Throat Infection
|
|
Nature of the sample
|
Number of Isolates
|
MAR
|
Number of Isolates
|
MAR
|
Number of Isolates
|
MAR
|
| Discharge |
112 |
50 |
28 |
8 |
13 |
7 |
| Tissue |
1 |
1 |
28 |
12 |
10 |
5 |
| Overall total |
113 |
51 (45.1%) |
56 |
20 (35.7%) |
23 |
12 (52.2%) |
Note. ENT: Ear, nose, and throat; MAR: Multiple antibiotics resistant.
The biofilm formation of multiple antibiotic-resistant bacterial isolates cultured from ENT infections is represented in Tables 5, 6, and Figure 2. Out of 113 isolates that were isolated from ear infections, 51 (45.1%) were MAR, and 36 (70.6%) were biofilm-producing organisms (BPOs). Further, from nose infection, there was a total of 46 isolates, including 20 (43.5%) MAR and 16 (80%) BPO. A total number of 23 isolates were recovered from the throat, including 13 (52.2%) MAR and 10 (83.3%) BPO. Of all collected samples (from both tissue and discharge samples), all the tissue samples that were multiple antibiotic resistant for gram-positive bacteria were 91.7% biofilm producers, and 100% of gram-negative bacteria were BPOs, while 71% and 61.8% were discharge samples and biofilm producers for both gram-positive and gram-negative, respectively.
Table 5.
Biofilm Formation of Multiple Antibiotics Resistant Isolate Cultured From Ear, Nose, and Throat Infection Samples
|
Gram-Negative Bacteria Isolate
|
Ear Infection
|
Nose Infection
|
Throat Infection
|
|
No. of Isolates (n=56)
|
No. of MAR (n=31)
|
No. of+ve BF (n=20)
|
No. of isolates (n=16)
|
No. of MAR (n=8)
|
No. of+ve BF (n=6)
|
No. of isolates (n=5)
|
No. of MAR (n=1)
|
No. of+ve BF (n=1)
|
|
Proteus spp. |
22 (39.2%) |
9 (29%) |
8 (40%) |
5 (31.2%) |
1 (12.5%) |
1 (16.7) |
2 (40%) |
0 |
0 |
|
Pseudomonas spp. |
15 (26.8%) |
13 (41.9%) |
7 (35%) |
4 (25%) |
3 (37.5%) |
2 (33.3) |
1 (20%) |
1 (100%) |
1 (100%) |
|
Escherichia coil
|
4 (7.1%) |
3 (9.7%) |
1 (5%) |
3 (18.8%) |
1 (12.5%) |
0 |
0 |
0 |
0 |
|
Klebsiella spp. |
4 (7.1%) |
2 (6.5%) |
2 (10%) |
1 (6.3%) |
1 (12.5%) |
1 (16.7) |
0 |
0 |
0 |
|
Citrobacter spp. |
5 (8.9%) |
2 (6.5%) |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Enterobacter spp. |
1 (1.8%) |
0 |
0 |
3 (18.8%) |
2 (25%) |
2 (33.3) |
0 |
0 |
0 |
|
Serratia spp. |
2 (3.6%) |
1 (3.2%) |
1 (5%) |
0 |
0 |
0 |
2 (40%) |
0 |
0 |
|
Salmonella spp. |
2 (3.6%) |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Acinetobacter sp. |
1 (1.8%) |
1 (3.2%) |
1 (5%) |
0 |
0 |
0 |
0 |
0 |
0 |
Note. MAR, multiple antibiotic resistant; BF: Biofilm.
Table 6.
Biofilm formation of Multiple Antibiotics Resistant Isolate Cultured From Ear, Nose, and Throat Infections Samples
|
Gram-Positive Bacteria Isolate
|
Ear Infection
|
Nose Infection
|
Throat Infection
|
|
No. of isolates (n=57)
|
No. of MAR (n=20)
|
No. of+ve BF (n=14)
|
No. of isolates (n=40)
|
No. of MAR (n=12)
|
No. of+ve BF (n=10)
|
No. of Isolates (n=18)
|
No. of MAR (n=11)
|
No. of+ve BF (n=9)
|
|
Staphylococcus aureus
|
19 (33.3%) |
9 (45%) |
9 (64.3%) |
15 (37.5%) |
8 (66.7%) |
7 (70%) |
7 (38.9%) |
6 (54.5%) |
4 (44.4%) |
|
Staphylococcus spp. |
11 (19.3%) |
4 (20%) |
4 (28.6%) |
8 (20%) |
3 (25%) |
2 (20%) |
2 (11.1%) |
1 (9.1%) |
1 (11.1%) |
|
Micrococcus spp. |
2 (3.5%) |
2 (10%) |
0 |
1 (2.5%) |
0 |
0 |
0 |
0 |
0 |
|
Enterococcus spp. |
1 (1.6%) |
0 |
0 |
2 (5%) |
1 (8.3%) |
1 (10%) |
0 |
0 |
0 |
|
Streptococcus spp. |
4 (7%) |
1 (5%) |
0 |
3 (7.5%) |
0 |
0 |
3 (16.7%) |
2 (18.2%) |
2 (22.2%) |
|
Corynebacterium spp. |
13 (22.8%) |
2 (10%) |
0 |
4 (10%) |
0 |
0 |
3 (16.7%) |
1 (9.1%) |
1 (11.1%) |
|
Bacillus spp. |
7 (12.3%) |
2 (10%) |
1 (7.1%) |
7 (17.5%) |
0 |
0 |
3 (16.7%) |
1 (9.1%) |
1 (11.1%) |
Note. BF: Biofilm; MAR: Multiple antibiotic resistance.
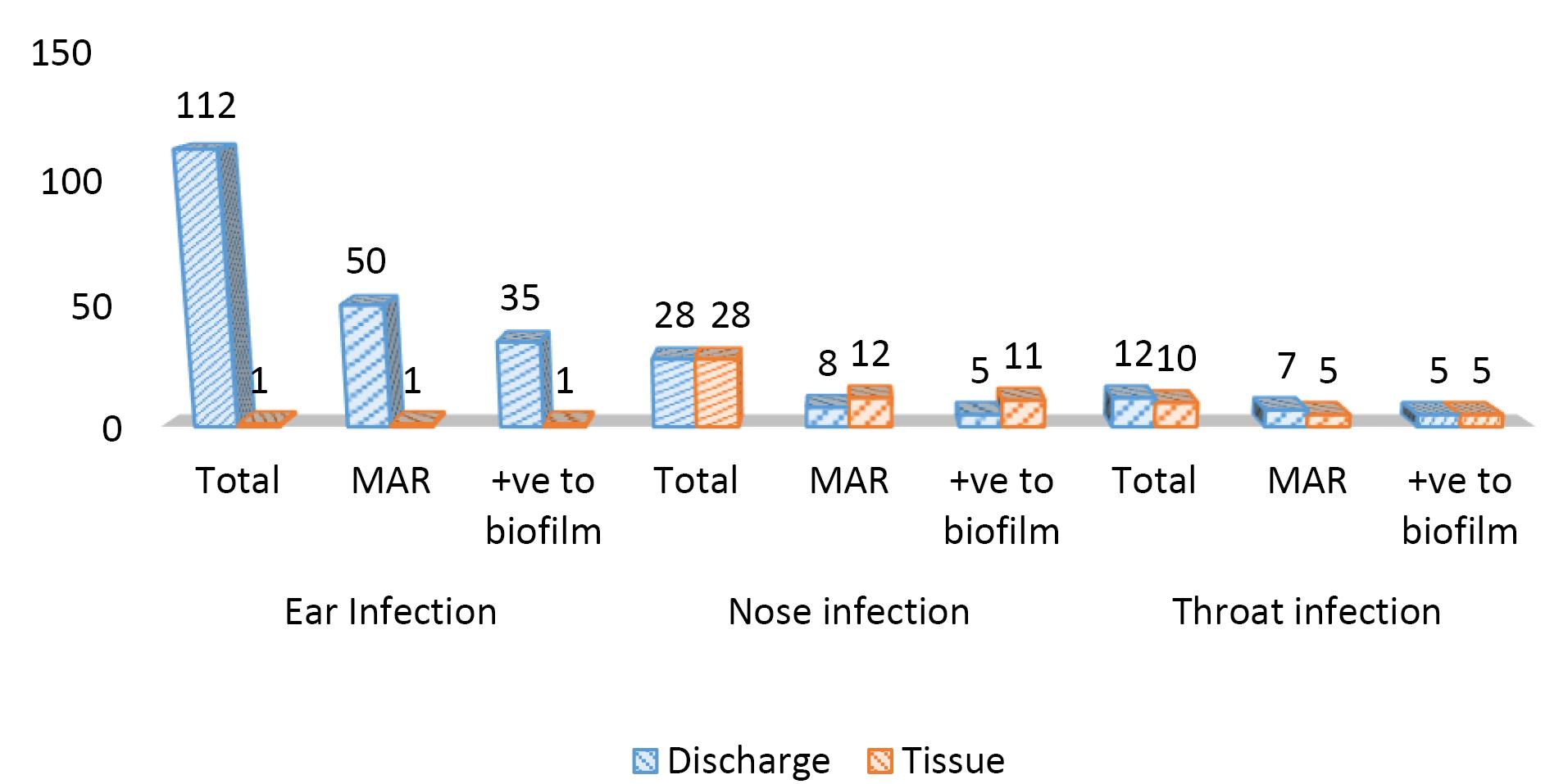
Figure 2.
The Frequency of Biofilm-Producing Bacteria Isolated From ENT Infections Related to Their Site. Note. ENT: Ear, nose, and throat
.
The Frequency of Biofilm-Producing Bacteria Isolated From ENT Infections Related to Their Site. Note. ENT: Ear, nose, and throat
Discussion
In this study, ear infections had the highest prevalence, followed by nasal and throat infections, which corroborates the findings of studies performed by Sharma et al (20) and Otoghile et al (21) in Guwahati, India and River State, Nigeria, respectively. Studies have identified different bacterial as aetiology agents causing ENT infections (22-26). In this study, the main isolated bacteria were S. aureus (21.4%), Proteus spp. (15.1%), Staphylococcus spp. (10.9%), Corynebacterium spp., and P. aeruginosa (10.4%), respectively. Similar to our reports, other investigators such as Obiajuru and Chukuezi (22) and Ahmad et al (24) implicated S. aureus as the most prevalent organism in Imo and Kano States in Nigeria, respectively. However, El-Mahmood et al (6) and Al-Badaii et al (26) reported Streptococcus pyogenes, S. aureus, Klebsiella pneumoniae, and P. aeruginosa as the commonest bacteria causing infections in Yola and Dhamar Governorate, Yemen, respectively. Generally, in this study, Gram-negative bacteria were most sensitive to meropenem and ofloxacin, while the Gram-positive bacteria were most sensitive to both vancomycin, meropenem, and ofloxacin, suggesting that these drugs may be adopted as empiric antibiotics. Heidari et al (27) also found P. aeruginosa as the main culprit, which also demonstrated high resistance to ciprofloxacin and amikacin. The observations showed that the isolates are highly resistant to beta-lactam, which is the most current empirical antibiotic used in our locality. The findings of previous usage of antibiotics commonly found among patients with drug resistance are not unexpected given the poor regulation of drug dispensing and usage in the Nigerian environment. The finding of this study underscores the need for periodic reviews of empiric antibiotics in our environment.
In this study, almost all the isolates, which are multiple antibiotic resistant, were positive for biofilm production, especially isolates from tissue cultures. S. aureus and P. aeruginosa isolates were able to produce biofilms in all the sites of ENT infection. The biofilm-producing bacteria have a high tendency to be more dominant in tissue samples compared to discharge samples, as found in this study. Their ability to stick to a surface or tissue, freely flow in the bloodstream, evade the immune system of the host, and resist the effect of appropriate drugs has been reported to play a major role in the persistence of bacterial infections. This ability has enabled them to avoid the possibility of being washed away through water flow or bloodstream, to oppose many bodily factors that can hamper the formation and effect of biofilm, and to tolerate any harsh environmental conditions (28-30). Therefore, this can enhance their potential to resist antibiotics. In other words, biofilm generation can make antibiotics lose their ability to fight bacterial infections by protecting bacteria strains from antibiotic agents and immune system cells, especially those who would have ordinarily been overpowered by appropriate medications (31), implying that the potentiality of a bacteria isolate to produce biofilm contributes significantly to their degree of antibiotic resistance. However, other researchers (5,28) agreed that there is a cordial relationship between AMR and the ability to form biofilm. They also perceived that the underlying mechanisms for this relationship can be influenced by the strain of bacteria, biofilm development level, concentration of extracellular polymeric substances in biofilm, their genetic regulatory system, and the type of antimicrobial agent.
Studies have reported that cases of chronic ENT infections, especially rhinosinusitis and otitis media, are mostly caused by the formation of bacterial biofilms on adenoids (9-12). These findings are in agreement with those of our study because almost all tissue isolates are MAR producing biofilms, including 100% for Gram-positive and 83.3% for Gram-negative organisms when compared with isolates from discharge samples. This information has not only exposed one of the major causes of MAR in ENT infection, but it has further explained the reasons for the magnitude of sequels and complications of ENT infections among our patients. While other factors, such as wrong use of antibiotics, improper infection prevention and control methods, and availability of antibiotics as over-the-counter medications, are also of public health importance, more intense focus should be given to combating BPOs (2). In many parts of the world, there has been progressively increasing in-depth insight into what is being understood about the mechanism of formation, structural integrity, effects of genetics, and clinical impacts of biofilms. This also includes the ability to understand the properties of biofilm-producing bacteria to overpower potent antimicrobial agents and the methods that can be employed to impair the potency of biofilm, such as probiotic-based intervention strategies (5,27,29). Therefore, solution-oriented local studies are needed to look more into available medicinal substances and potent anti-biofilm agents that can militate against biofilms in an attempt to disarm the effect of MAR in the management of ENT infections.
Conclusion
The ability of common bacteria causing ENT infections to resist antibiotic agents is enormous and calls for urgent attention. BPOs are mostly common among the causative agents for ENT infections. These biofilm-producing bacteria have higher tendencies to dominate in body-infected tissues other than the discharges being produced; accordingly, tissue biopsy for culture and sensitivity should be considered more appropriate where visible, particularly when confronted with hard-to-treat infections in ENT clinical settings. The proper sensitivity pattern of the isolate must be effectively investigated before prescribing antibiotics to reduce the prevalence of ENT infection. There is also an urgent need to consider anti-biofilm antibiotic agents with the development of standardized anti-biofilm protocols in the prevention and management of multiple antibiotic resistance.
Authors’ Contribution
Conceptualization: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Anthonia Olufunke Oluduro.
Data curation: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Akumbu Sylvia Nwamuo.
Formal analysis: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele.
Investigation: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele, Akumbu Sylvia Nwamuo.
Methodology: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Anthonia Olufunke Oluduro.
Project administration: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele, Akumbu Sylvia Nwamuo, Anthonia Olufunke Oluduro.
Resources: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele, Akumbu Sylvia Nwamuo, Anthonia Olufunke Oluduro.
Software: Oluwabusayomi Roseline Ademakinwa, Samuel Oluyomi Ayodele.
Supervision: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Anthonia Olufunke Oluduro.
Validation: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele, Akumbu Sylvia Nwamuo, Anthonia Olufunke Oluduro.
Visualization: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele, Akumbu Sylvia Nwamuo, Anthonia Olufunke Oluduro.
Writing–original draft: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo.
Writing–review & editing: Oluwabusayomi Roseline Ademakinwa, Adekunle Adeyemo, Samuel Oluyomi Ayodele, Anthonia Olufunke Oluduro.
Competing Interests
The authors declare that they have no conflict of interests.
Ethical Approval
This study was performed in line with the principles of the Declaration of Helsinki. Ethical approval was also granted by the Ethical and Research Committee of the Obafemi Awolowo University Teaching Hospitals Complex, Ile-Ife, Nigeria (with protocol number: ERC/2018/06/14) before commencement of the study.
Funding
We declare that no funds, grants, or any other support were received during the data collection and preparation of this manuscript.
References
- World Health Organization (WHO). Antimicrobial Resistance. WHO; 2014. p. 2-8.
- Dahal A, Shrestha K, Karki R, Bhattarai S, Aryal S, Deo SK. Antimicrobial resistance and biofilm production in uropathogens from renal disease patients admitted to Tribhuvan University Teaching Hospital, Nepal. J Clin Pharm Ther 2023; 2023(1):4867817. doi: 10.1155/2023/4867817 [Crossref] [ Google Scholar]
- Gorems K, Beyene G, Berhane M, Mekonnen Z. Antimicrobial susceptibility patterns of bacteria isolated from patients with ear discharge in Jimma town, southwest, Ethiopia. BMC Ear Nose Throat Disord 2018; 18:17. doi: 10.1186/s12901-018-0065-0 [Crossref] [ Google Scholar]
- Solanki P, Sultana Y, Singh S. Traditional medicine: exploring their potential in overcoming multi-drug resistance. In: Aggarwal G, ed. Strategies to Overcome Superbug Invasions: Emerging Research and Opportunities. 1st ed. Pennsylvania, USA: IGI Global; 2021. p. 118-29.
- Sharan M, Dhaka P, Bedi JS, Mehta N, Singh R. Assessment of biofilm-forming capacity and multidrug resistance in Staphylococcus aureus isolates from animal-source foods: implications for lactic acid bacteria intervention. Ann Microbiol 2024; 74(1):22. doi: 10.1186/s13213-024-01768-5 [Crossref] [ Google Scholar]
- El-Mahmood AM, Isa H, Mohammed A, Tirmidhi AB. Antimicrobial susceptibility of some respiratory tract pathogens to commonly used antibiotics at the specialist hospital, Yola, Adamawa state, Nigeria. J Clin Med Res 2010; 2(8):135-42. [ Google Scholar]
- Kabeer S, Zafar A, Mehdi N, Zubair M, Javed H, Shaheen AJ. Isolation and antimicrobial susceptible pattern of bacterial pathogens from ear, nose and throat of paediatric patients. Pak J Med Health Sci 2014; 8(3):644-7. [ Google Scholar]
-
Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr 2016;4(2). doi: 10.1128/microbiolspec.VMBF-0016-2015.
- Smith A, Buchinsky FJ, Post JC. Eradicating chronic ear, nose, and throat infections: a systematically conducted literature review of advances in biofilm treatment. Otolaryngol Head Neck Surg 2011; 144(3):338-47. doi: 10.1177/0194599810391620 [Crossref] [ Google Scholar]
- Coticchia J, Zuliani G, Coleman C, Carron M, Gurrola J 2nd, Haupert M. Biofilm surface area in the pediatric nasopharynx: chronic rhinosinusitis vs obstructive sleep apnea. Arch Otolaryngol Head Neck Surg 2007; 133(2):110-4. doi: 10.1001/archotol.133.2.110 [Crossref] [ Google Scholar]
- Marseglia GL, Caimmi D, Pagella F, Matti E, Labó E, Licari A. Adenoids during childhood: the facts. Int J Immunopathol Pharmacol 2011; 24(4 Suppl):1-5. doi: 10.1177/03946320110240s401 [Crossref] [ Google Scholar]
- Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 2002; 287(13):1710-5. doi: 10.1001/jama.287.13.1710 [Crossref] [ Google Scholar]
- Drago L, Pignataro L, Torretta S. Microbiological aspects of acute and chronic pediatric rhinosinusitis. J Clin Med 2019; 8(2):149. doi: 10.3390/jcm8020149 [Crossref] [ Google Scholar]
- Bezerra TF, Padua FG, Gebrim EM, Saldiva PH, Voegels RL. Biofilms in chronic rhinosinusitis with nasal polyps. Otolaryngol Head Neck Surg 2011; 144(4):612-6. doi: 10.1177/0194599811399536 [Crossref] [ Google Scholar]
- Tan NC, Tran HB, Foreman A, Jardeleza C, Vreugde S, Wormald PJ. Identifying intracellular Staphylococcus aureus in chronic rhinosinusitis: a direct comparison of techniques. Am J Rhinol Allergy 2012; 26(6):444-9. doi: 10.2500/ajra.2012.26.3822 [Crossref] [ Google Scholar]
- Holt JG, Krieg NR, Sneath PH, Staley JT, Williams T, Hensyl WR. Bergey’s Manual of Determinative Bacteriology. 9th ed. Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Maryland: Williams and Wilkins Co Baltimore; 1994. p. 151-7.
- Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 1966; 45(4):493-6. [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI Supplement M100. Pennsylvania: CLSI; 2017.
- Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis 2011; 15(4):305-11. doi: 10.1590/s1413-86702011000400002 [Crossref] [ Google Scholar]
- Sharma K, Bhattacharjya D, Barman H, Goswami SC. Common ear, nose, and throat problems in pediatric age group presenting to the emergency clinic prevalence and management: a hospital-based study. Indian J Clin Pract 2014; 24(8):756-60. [ Google Scholar]
- Otoghile B, Ibekwe MU, Totyen EL, Inei AN. Attitude of parents in a Niger Delta community towards ear, nose and throat diseases among their children: a preliminary study Port Harcourt Medical Journal. 2019 S ep 1; 13(3):98-101. doi: 10.4103/phmj.phmj_19_19 [Crossref] [ Google Scholar]
- Obiajuru IO, Chukuezi AB. Microbiological assessment of ENT infections and diseases: clinical case study at Orlu, Imo state, Nigeria. J Res Nurs Midwifery 2013; 2(5):72-6. [ Google Scholar]
- Kumar R, Srivastava P, Sharma M, Rishi S, Nirwan PS, Hemwani K. Isolation and antimicrobial sensitivity profile of bacterial agents in chronic suppurative otitis media patients at NIMS Hospital Jaipur. Int J Pharm Biol Sci 2013; 3(4):265-9. [ Google Scholar]
- Ahmad MM, Kurawa ZM, Shu’aibu I, Yahaya G. Microbiological assessment of bacterial isolates from ear, nose and throat (ENT) among patients attending Aminu Kano Teaching Hospital. Niger J Basic Appl Sci 2016; 24(1):15-8. doi: 10.4314/njbas.v24i1.3 [Crossref] [ Google Scholar]
- Phukan S, Das S. A study on the prescribing pattern of antimicobial drugs in patients attending the ear, nose, throat department of a tertiary care teaching hospital. Asian J Pharm Clin Res 2020; 13(6):197-9. doi: 10.22159/ajpcr.2020.v13i6.37223 [Crossref] [ Google Scholar]
- Al-Badaii F, Al-Taibi A, Al-Shaeri H, Homied E, Obad M, Al-Khatari F. Isolation, identification and antibiotic susceptibility of bacteria from upper respiratory tract infections at Dhamar governorate, Yemen. Int J Sci Res Biol Sci 2021; 8(2):12-9. [ Google Scholar]
- Heidari R, Farajzadeh Sheikh A, Hashemzadeh M, Farshadzadeh Z, Salmanzadeh S, Saki M. Antibiotic resistance, biofilm production ability and genetic diversity of carbapenem-resistant Pseudomonas aeruginosa strains isolated from nosocomial infections in southwestern Iran. Mol Biol Rep 2022; 49(5):3811-22. doi: 10.1007/s11033-022-07225-3 [Crossref] [ Google Scholar]
- Zamani S, Mohammadi A, Hajikhani B, Abiri P, Fazeli M, Nasiri MJ. Mupirocin-resistant Staphylococcus aureus in Iran: a biofilm production and genetic characteristics. Biomed Res Int 2022; 2022:7408029. doi: 10.1155/2022/7408029 [Crossref] [ Google Scholar]
- Rather MA, Gupta K, Mandal M. Microbial biofilm: formation, architecture, antibiotic resistance, and control strategies. Braz J Microbiol 2021; 52(4):1701-18. doi: 10.1007/s42770-021-00624-x [Crossref] [ Google Scholar]
- Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem 2015; 7(4):493-512. doi: 10.4155/fmc.15.6 [Crossref] [ Google Scholar]
- Akrami S, Ekrami A, Avarvand AY. Biofilm generation and antibiotic resistant profile of extensive and multidrug resistant Pseudomonas aeruginosa from burn patients in Ahvaz: a cross-sectional study. Health Sci Rep 2024; 7(6):e2138. doi: 10.1002/hsr2.2138 [Crossref] [ Google Scholar]