Avicenna Journal of Clinical Microbiology and Infection. 11(1):1-8.
doi: 10.34172/ajcmi.3509
Original Article
Alterations in the Gut Microbiota Composition and Structure of Children Under 5 Years With Sepsis: A Case Report From a Federal Medical Center in Lagos State, Nigeria
Tenny Obiageli Gladys Egwuatu 1, *  , Uzoma Nonyelum Okeke 1, Sakinat Bello 1, Awosipe Ayobola Oluwaseun 1, Abraham Ajayi 2
, Uzoma Nonyelum Okeke 1, Sakinat Bello 1, Awosipe Ayobola Oluwaseun 1, Abraham Ajayi 2  , Utibeima Udo Essiet 2
, Utibeima Udo Essiet 2  , Agatha David 3
, Agatha David 3  , Nnenna Kalu 4, Afolabi Lesi 5, Stella Ifeanyi Smith 2, 6
, Nnenna Kalu 4, Afolabi Lesi 5, Stella Ifeanyi Smith 2, 6 
Author information:
1Department of Microbiology, University of Lagos Akoka, Nigeria
2Molecular Biology and Biotechnology Department, Nigerian Institute of Medical Research, Yaba, Lagos, Nigeria
3Clinical Sciences Department, Nigerian Institute of Medical Research, Yaba, Lagos, Nigeria
4Federal Medical Center Ebute-Metta, Lagos Mainland, Ebute-Metta, Lagos State, Nigeria
5College of Medicine, University of Lagos Idi-Araba, Lagos Nigeria
6Department of Biological Sciences, Mountain Top University, Ogun State, Nigeria
Abstract
Background: Sepsis is a major cause of death among children in Sub-Saharan Africa. Although studies have suggested that the dysbiosis of the gut microbiota is associated with sepsis, there are limited data on the microbiota profile of children with sepsis in low- and middle-income countries such as Nigeria.
Methods: To address this gap, this study was conducted to examine the gut microbiota of four children (S2, S4, S6, and S8) with sepsis and two apparently healthy controls (C2 and C4). DNA was extracted from stool samples, and 16S rRNA amplicon sequencing and bioinformatics analysis were used to determine the microbial composition of the microbiota.
Results: A significant reduction in the abundance of the phylum Bacteroidetes was observed in sepsis cases, while the family Enterobacteriaceae was dominant, with a relative abundance of 29.7%, 36.9%, 47.4%, and 34.6% in S2, S4, S6, and S8, respectively. Pathogenic bacterial species such as Megasphaera spp., Sutterella spp., and Clostridium difficile were dominant among some of the sepsis samples. The bacterial communities of the microbiota of participants with sepsis largely diverged from those of the controls, with a remarkable decrease in diversity.
Conclusion: This study highlights an alteration in the gut microbiota of children presenting with sepsis. Notably, there was a reduction in beneficial commensals and an increase in potential pathogenic bacterial species. Since sepsis remains a major public health challenge, there is an urgent need to explore the gut microbiota for possible intervention.
Keywords: Bacteria, Children, Gut microbiota, Metagenomics, Sepsis
Copyright and License Information
© 2024 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Egwuatu TOG, Okeke UN, Bello S, Oluwaseun AA, Ajayi A, Essiet UU, David A, Kalu N, Lesi A, Smith SI. Alterations in the gut microbiota composition and structure of children under 5 years with sepsis: a case report from a federal medical center in Lagos State, Nigeria. Avicenna J Clin Microbiol Infect. 2024; 11(1):1-8. doi:10.34172/ajcmi.3509
Introduction
The human gut microbiota plays a crucial role in human health and disease (1). It is involved in regulating the immune system, harnessing energy, and maintaining intestinal function (2). The diversity and structure of the gut microbiota serve as indicators for understanding behavioral patterns, evolutionary biology pathways, and potential pathologies in both humans and animals (3,4). Alterations in the community composition and metabolic products of the gut microbiota are associated with various diseases and health conditions, including autism spectrum disorders, neurodegenerative disorders, obesity, type 2 diabetes, cardiovascular disease, cancers, and inflammatory bowel disease.
For example, Zhuang et al (5) observed a reduction in the abundance of Bacteroidetes and an increased abundance of Actinobacteria among patients with autism spectrum disorders in China. Another study by Kang et al (6) demonstrated improved gastrointestinal and autism symptoms resulting from microbiota transfer therapy that enhanced bacterial diversity with increased populations of Bifidobacterium, Prevotella, and Desulfovibrio, among other taxa, in the gut of patients with autism spectrum disorders.
These findings underscore the significance of the gut microbiota in various health conditions and the potential for microbiota-based interventions to address these conditions. The gut microbiota has been implicated in various disorders and disease conditions, including sepsis. During sepsis, the microbiome undergoes significant disturbances, characterized by a sharp decrease in diversity, the loss of commensal bacteria, and the overgrowth of potential pathogenic bacteria such as Enterococcus and Staphylococcus (7). This disruption of the gut microbial communities has been associated with susceptibility to sepsis and potentially severe consequences (8). Sepsis, a life-threatening multiple-organ dysfunction caused by a dysregulated host response to infection, is a leading cause of morbidity and mortality in children globally, particularly in resource-limited settings such as Africa (9-11). In Nigeria, the burden of sepsis is substantial, with an estimated 16% of special care baby unit admissions in a tertiary hospital in southwest Nigeria attributed to sepsis (12). The gut microbiota’s role in sepsis and its potential as an auxiliary prognostic marker for sepsis in children have been the focus of recent research, highlighting the importance of understanding and managing the gut microbiota in the context of sepsis.
These findings suggest that sepsis is a significant health concern in Nigeria that necessitates comprehensive approaches to address it. One potential approach is to modulate the gut microbiota. To achieve this, it is crucial to understand the microbial composition of the gut microbiota in sepsis cases at a local level, as factors such as diet, host genetics, and environmental conditions influence the gut microbiota composition of hosts (13). Consequently, this study investigated the gut microbiota of children presenting with sepsis in Lagos, Nigeria, and compared it with the microbiota of apparently healthy children.
Materials and Methods
Study Design and Study Area
This cross-sectional study, involving four children with sepsis and two controls, was conducted at the Pediatric Unit of the Federal Medical Center (FMC), Ebute-Metta, Lagos. FMC Ebute-Metta is a 200-bed tertiary health facility with a vibrant pediatric unit and a fully functional intensive care unit. It is located on mainland Lagos and provides care to people of all socio-economic strata from around the Ebute-Metta, Bariga, Ojuelegba, Shomolu, and Yaba axes of Lagos State.
In this study, the cases were participants aged less than five years and presumed to have sepsis based on the criteria for defining sepsis in low- and middle-income countries previously described by Nwankwor et al (11), including symptoms not limited to fever, hypothermia, lethargy, abnormal heart rate for age (tachycardia or bradycardia), and irritability. The controls were apparently healthy children in the same age bracket of less than five years. Other inclusion criteria were the non-use of antibiotic therapy in the present illness before enrolment in the study (for the cases) and written guardian informed consent for children/wards to participate in the study. Information, including age, gender, weight, signs, and symptoms of sepsis, was collected by the study pediatrician using a pre-tested structured study questionnaire.
Sample Collection, DNA Extraction, and Sequencing
Six fecal samples were collected from four participants (S2, S4, S6, and S8) presenting with sepsis and two apparently healthy participants (Control 1 and Control 2). The samples were collected in sterile universal bottles and transported in a Thermobox at 4°C to the laboratory. Stool samples were then kept at -80 °C until DNA extraction. Total community DNA (tcDNA) was extracted using the QIAamp DNA Stool Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. The concentration and purity of tcDNA were determined by a NanoDrop 2000 UV spectrophotometer (Thermo Scientific, Wilmington, USA). The quality of tcDNA was determined by 0.8% agarose gel electrophoresis stained with ethidium bromide and visualized with a Clear View UV transilluminator fitted with a photo-documentation system (Clever Scientific Ltd., England) as previously described by Oyetibo et al (14). The extracted tcDNA was stored at -20°C for further use. In addition, 1 µL of aliquots from each tcDNA extract was used for the generation of libraries. The libraries were created with primers targeting the hypervariable region V3-V4 of the bacterial 16S rRNA gene using the primer sets 341F (5’-ACT CCT ACG GGA GGC AGC AG-3’) and 806R (5’-GGA CTACHV GGG TWT CTA AT-3’). Sequencing was performed on the Illumina HiSeq 2500 platform (Illumina San Diego, CA, USA) (15).
Data Analysis
Quality checks on raw FASTQ illumine sequence reads were performed using FastQC software, version 0.11.5 (16). The sequences of low quality were removed, and the remaining reads were assigned to operational taxonomic units with a threshold cut-off value of 97% using the quantitative insight into microbial ecology V.2.0 (QIIME 2) software (5). The diversity of the microbiota community of the samples was calculated based on their operational taxonomic unit profile using the R package vegan. The generated raw sequence reads were deposited in the sequence read archive database of the National Center for Biotechnology Information under BioProject with the accession number PRJNA1000308.
Results
Characteristics of Sepsis
Table 1 provides some characteristics of the participants who were involved in the study. The ages of the participants ranged from 2 weeks to 3 years. For the cases of sepsis, the body temperature ranged from 38.6 °C to 39.8 °C, and the average duration of fever before admission was 4.5 days. The diet of the participants aged 2 weeks and 3 months was breast milk only, while that of the participants aged 1.2 years was breast milk and other solid foods. One of the four participants died after 12 days of admission.
Table 1.
Baseline Clinical and Laboratory Characteristics of Study Participants With Sepsis
|
Code
|
Gender
|
Age
|
Temperature at Presentation (°C)
|
Duration of Fever
|
Diet
|
WBC
|
Fatality
|
| S2 |
Male |
2 months |
39.1 |
7 days |
Breast milk |
22500 |
Yes |
| S4 |
Female |
2 weeks |
39.0 |
2 days |
Breast milk |
18800 |
No |
| S6 |
Male |
1.6 years |
39.8 |
5 days |
Breast milk and other foods |
18500 |
No |
| S8 |
Male |
1.2 years |
38.6 |
4 days |
Breast milk and other foods |
15000 |
No |
| C2 |
Male |
3 years |
36.6 |
NA |
Family diet |
ND |
NA |
| C4 |
Male |
3 years |
36.9 |
NA |
Family diet |
ND |
NA |
Note. WBC: White blood cell count; NA: Not applicable; ND: Not determined.
Phylum
The microbial compositions of the gut of participants with sepsis (S2, S4, S6, and S8) and apparently healthy controls analyzed at various taxonomic levels revealed that Firmicutes was the dominant phyla across all samples, except for S4, which had an 11.0% relative abundance of Firmicutes. Bacteroidetes had a relative abundance of 25.0% and 43.5% in control 1 and control 2, respectively, while a sharp decrease of Bacteroidetes was observed in S8 (12.5%), S6 (0.08%), S4 (0.03%), and S2 (0.4%). Proteobacteria were dominant in S2, S4, S6, and S8, with a relative abundance of 29.9%, 49.0%, 48.7%, and 34.9%, respectively (Figure 1). S4 had the highest (39.3%) abundance of Actinobacteria, followed by S2 (25.6%) and S8 (14.9%). The phylum Fusobacteria was only present in control 2, with a relative abundance of 0.5%.
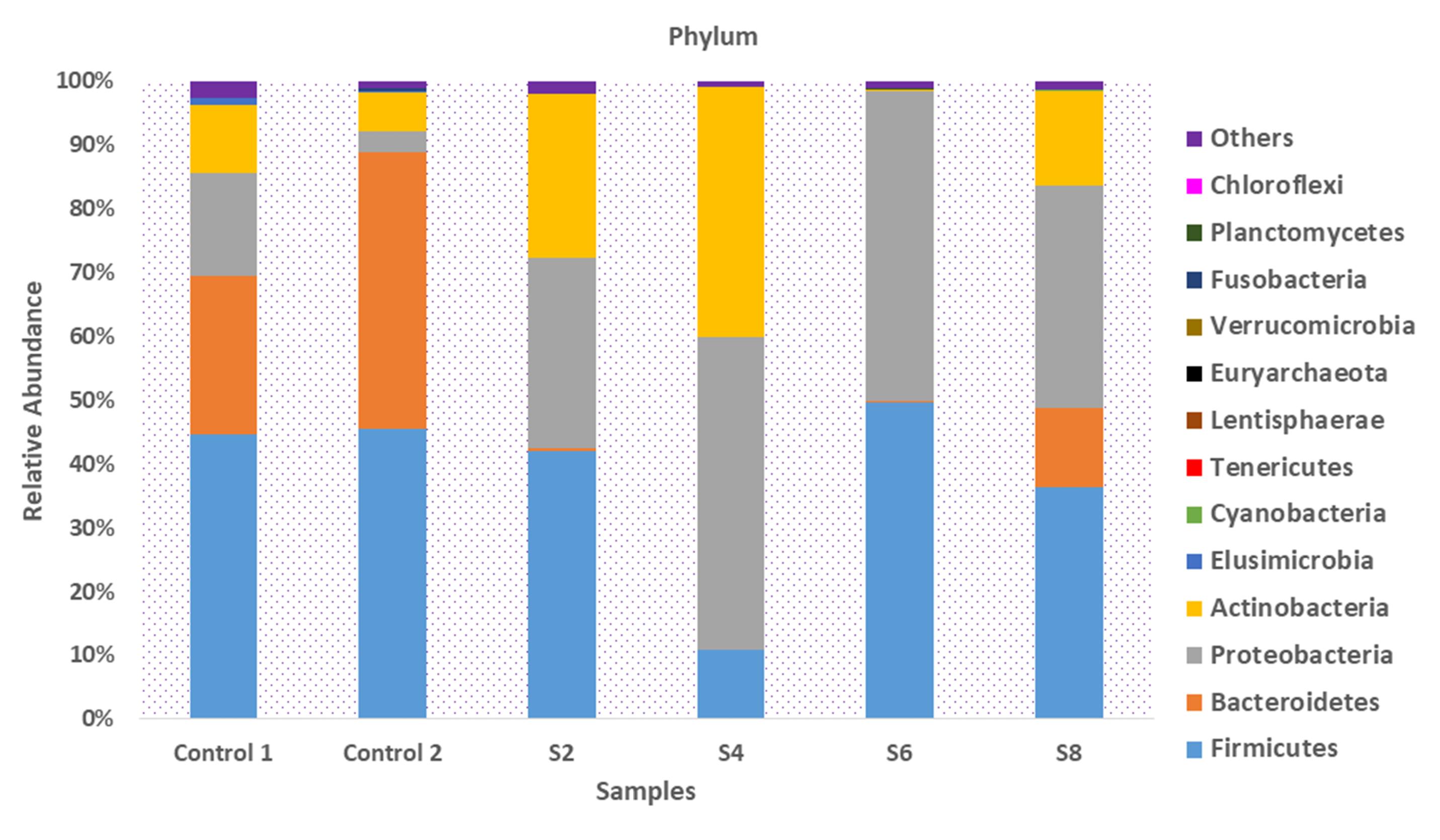
Figure 1.
Taxonomic Composition of the Microbiota of Participants Presenting With Sepsis and Apparently Healthy Controls at the Phylum Level
.
Taxonomic Composition of the Microbiota of Participants Presenting With Sepsis and Apparently Healthy Controls at the Phylum Level
Family
At the family level, Bacteroidaceae were absent in S2, S4, and S6. However, there was a relative abundance of Bacteroidaceae (12.1%) in S8. The relative abundance of Veillonellaceae observed across samples was 7.1%, 11.2%, 27.1%, 0.01%, and 11.9% in Control 1, Control 2, S2, S6, and S8, respectively. The family Bifidobacteriaceae was dominant in S2 (25.4%) and S4 (39.1%), while it was relatively low in S6 (0.1%) and S8 (7.2%). Enterobacteriaceae was dominant across all samples with sepsis, with a relative abundance of 29.7%, 36.9%, 47.4%, and 34.6% in S2, S4, S6, and S8, respectively (Figure 2). On the other hand, control 1 and control 2 had a limited presence of Enterobacteriaceae, with a relative abundance of 2.0% and 0.1%, respectively.
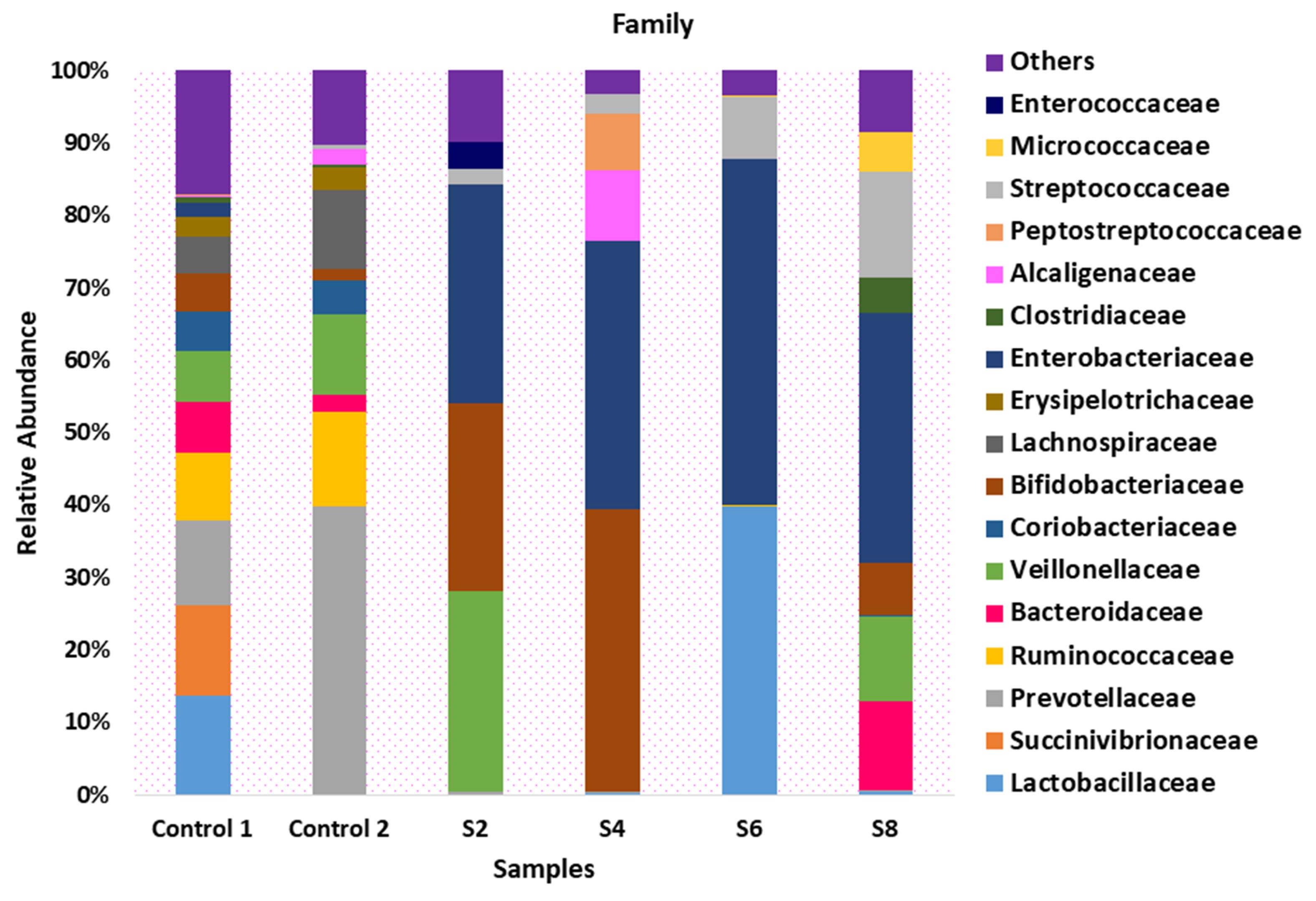
Figure 2.
Taxonomic Composition of the Microbiota of Participants Presenting With Sepsis and Apparently Healthy Controls at the Family Level
.
Taxonomic Composition of the Microbiota of Participants Presenting With Sepsis and Apparently Healthy Controls at the Family Level
Genus
At the genus level, the dominant genus in the control samples was Prevotella. Bifidobacterium was the dominant genus in S2 and S4, with a relative abundance of 25.4% and 39.1%, respectively. Bacteroides were dominant in S8 (12.1%), and there was a relative abundance of Streptococcus in the microbiota of all four participants with sepsis (Figure 3). The relative abundance of microbiota sequences revealed that the microbial structure of the control group differed from that of the sepsis group.
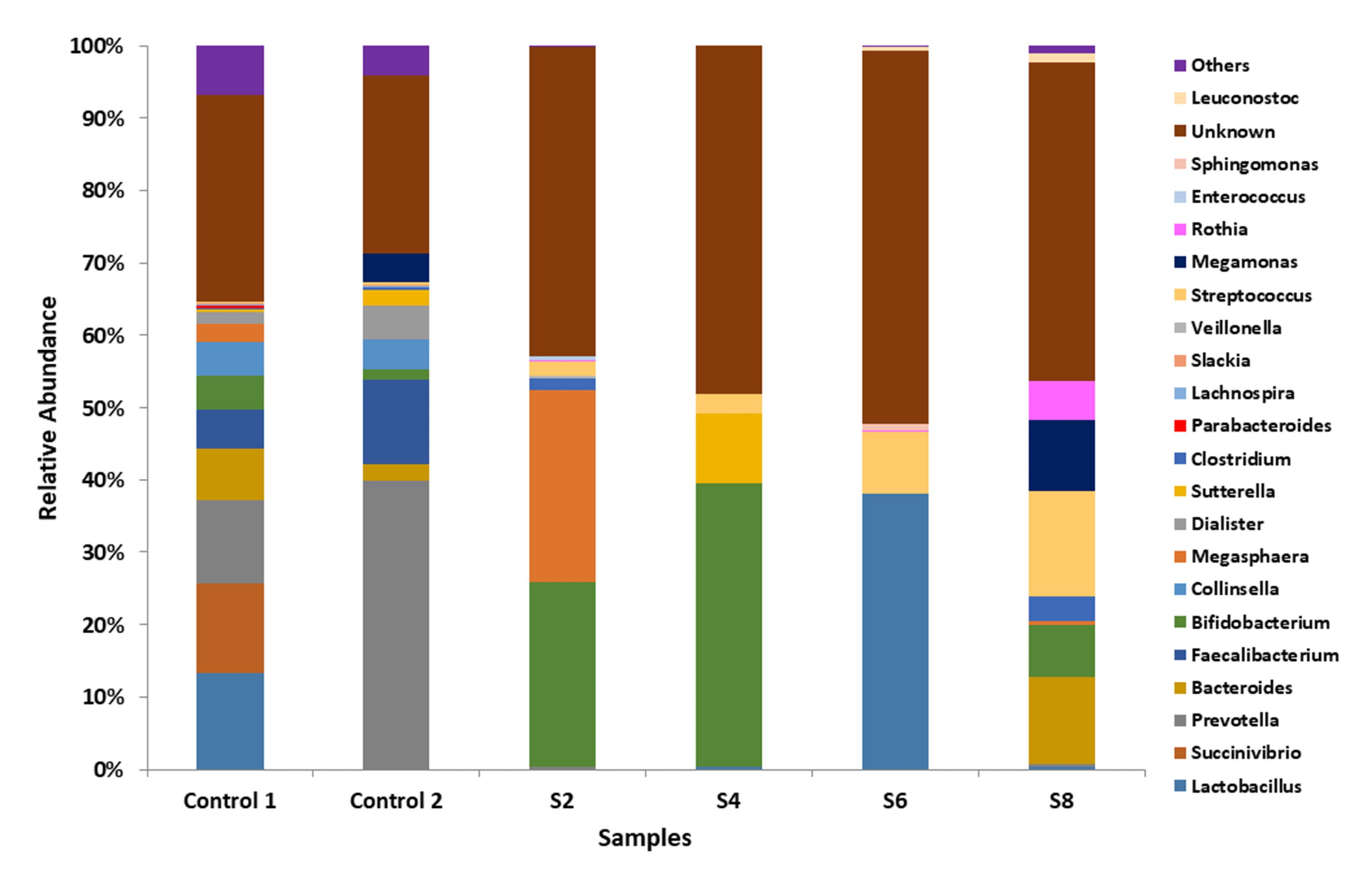
Figure 3.
Taxonomic Composition of the Microbiota of Participants Presenting With Sepsis and Apparently Healthy Controls at the Genus Level
.
Taxonomic Composition of the Microbiota of Participants Presenting With Sepsis and Apparently Healthy Controls at the Genus Level
Species
Figures 4A, 4B and 4C illustrate the percentage relative abundance of top species present in the microbiota of participants presenting with sepsis and apparently healthy controls. Lactobacillus ruminis and Succinivrio spp. were the dominant species in Control 1, while Prevotella copri and Prevotella stercorea were the dominant species in Control 2 (Figure 4A). Megasphaera spp. and Bifidobacterium bifidum, as well as Sutterella spp. and Bifidobacterium breve, were dominant species in S2 and S4, respectively. Moreover, Lactobacillus spp. and Streptococcus spp., as well as Bacteroides spp. and Megamonas spp., were dominant species in S6 and S8, respectively (Figure 4B). Streptococcus equi had a relative abundance of 0.04%, 0.08%, and 0.1% in S4, S6, and S8, respectively. However, S. equi was low or non-existent in S2. Clostridioides difficile was another dominant species in S4, with a relative abundance of 0.03%.
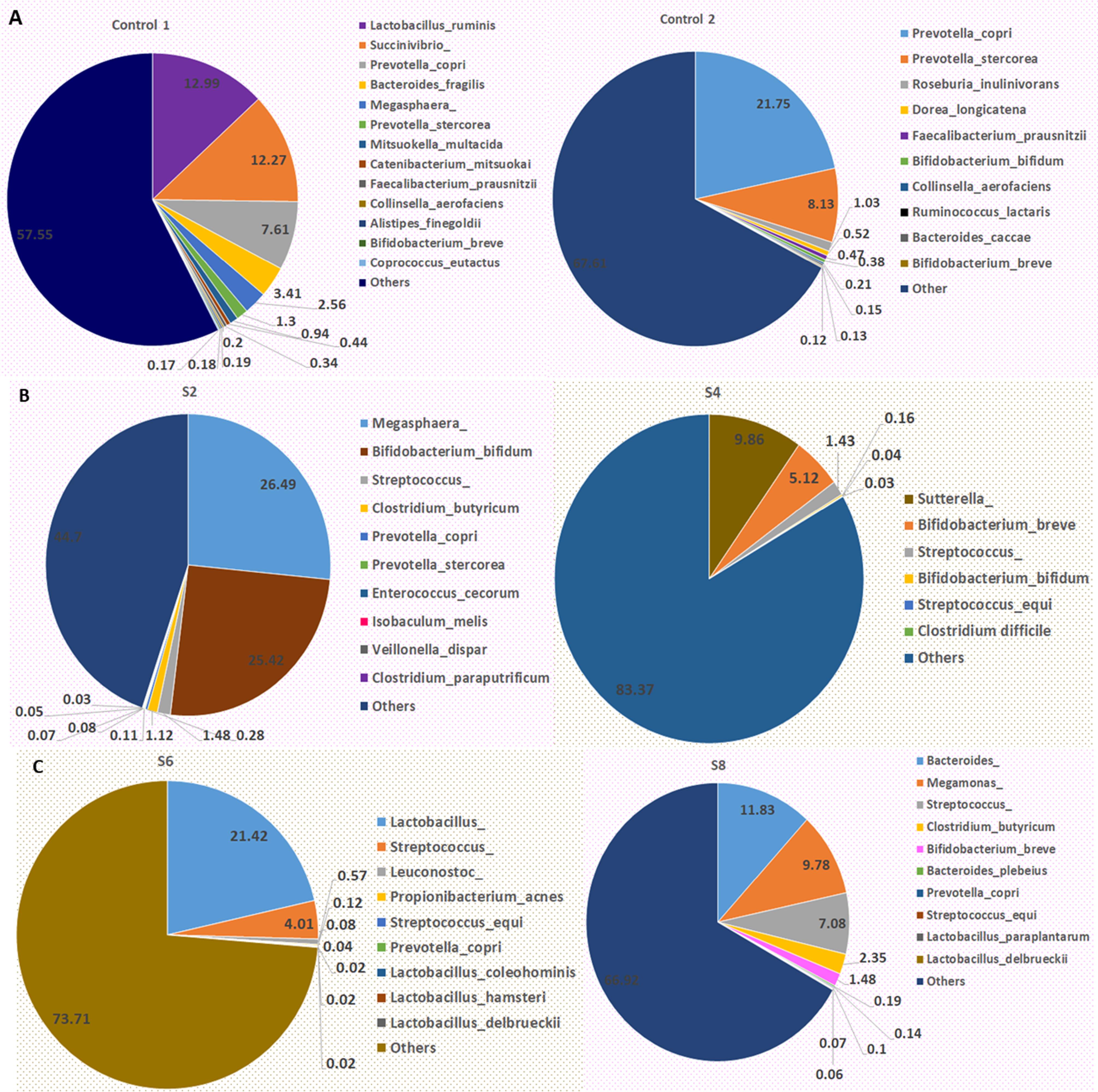
Figure 4.
Bacterial Species Abundance in the Microbiota of Participants: (A) Relative Abundance of Species in Control 1 and Control 2, (B) and (C) Relative Abundance of Bacterial Species in S2, S4, S6, and S8
.
Bacterial Species Abundance in the Microbiota of Participants: (A) Relative Abundance of Species in Control 1 and Control 2, (B) and (C) Relative Abundance of Bacterial Species in S2, S4, S6, and S8
Alpha and Beta Diversities
The mean community diversity indexes, including Shannon and Simpson (Table 2), revealed that the richness of the bacterial community in participants with sepsis was relatively low. Although the bacterial read counts of participants with sepsis in some cases (S4: 174,067; S8: 207,543) exceeded those of control participants, the gut microbiota of the controls was more diverse. The bacterial community of control samples was clustered with tight overlap, indicating the presence of similar bacterial populations. However, the bacterial communities of the microbiota of participants with sepsis largely diverged from those of the controls (Figure 5).
Table 2.
Alpha Diversity of Microbial Community Indicating Evenness, Richness, and Varieties of Species in Samples
|
|
Bacterial Reads Count
|
Chao1
|
ACE
|
Shannon
|
Simpson
|
Invsimpson
|
| C2 |
184,947 |
13 |
15 |
0.8 |
0.38 |
1.22 |
| C4 |
166,330 |
23 |
43 |
2.18 |
0.87 |
7.75 |
| S2 |
148,441 |
8 |
10 |
1.38 |
0.72 |
2.83 |
| S4 |
174,067 |
9 |
8 |
1.66 |
0.77 |
4.32 |
| S6 |
106,564 |
15 |
15 |
2.08 |
0.63 |
5.38 |
| S8 |
207,543 |
21 |
20 |
2.63 |
0.89 |
9.10 |
Note. ACE: Abundance-based Coverage Estimator
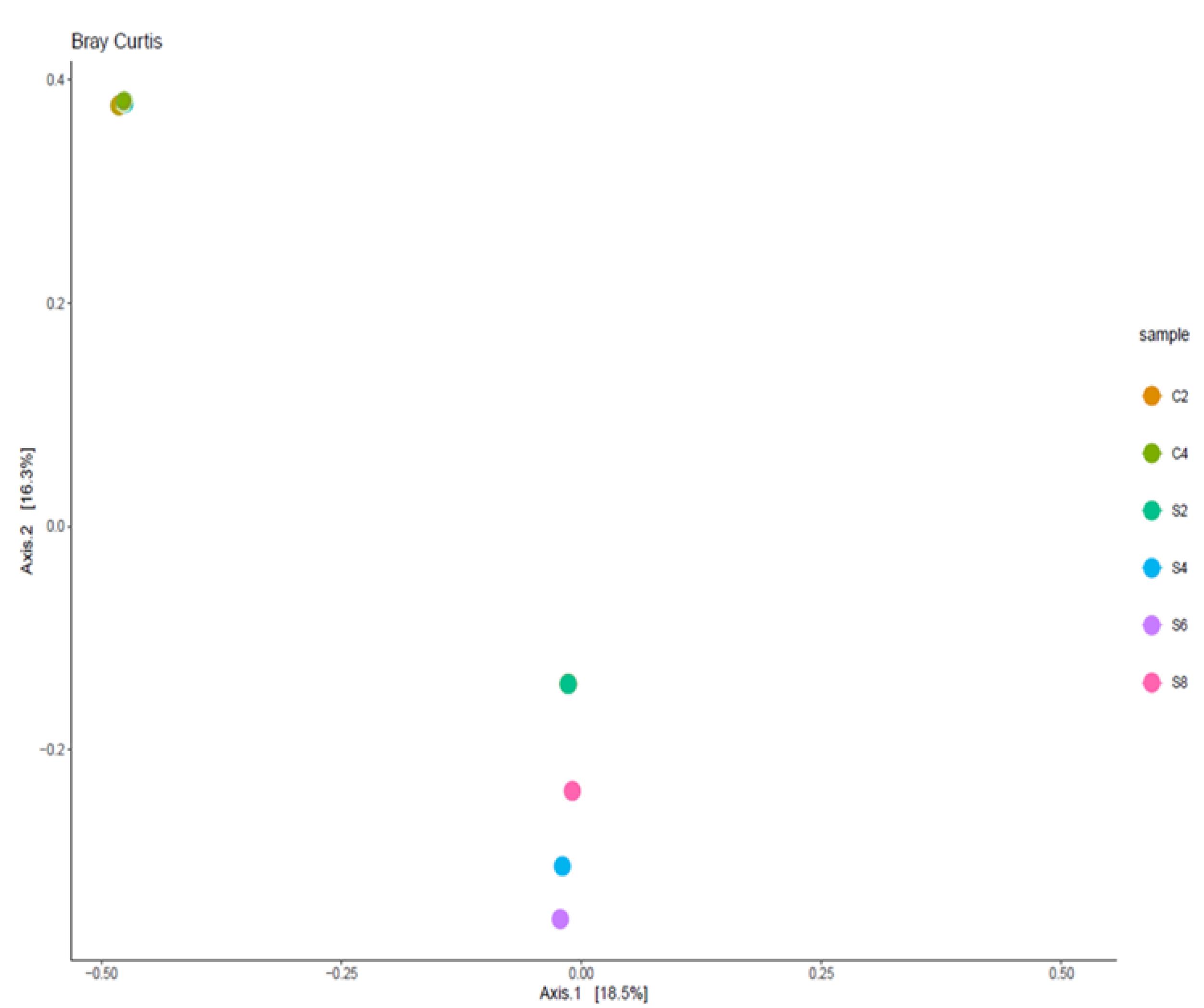
Figure 5.
Principal Coordinate Analysis Plot Showing Beta Diversity of Bacterial Communities of Control and Sepsis Participants Note. The bacterial communities of the controls demonstrate remarkable overlapping while those of participants with sepsis spread out
.
Principal Coordinate Analysis Plot Showing Beta Diversity of Bacterial Communities of Control and Sepsis Participants Note. The bacterial communities of the controls demonstrate remarkable overlapping while those of participants with sepsis spread out
Discussion
In this study, participants with sepsis presented with body temperatures ranging from 38.6 °C to 39.8 °C and an average of 7 days of onset of fever, which correlates with the elevated body temperature of defined sepsis cases according to the World Health Organization (11). However, the prolonged duration of the onset of fever before admission at the FMC in some of the sepsis cases could be attributed to the low economic status of the parents of the participants, who might have visited other health facilities with poor or inadequate personnel and were referred to the FMC when their conditions were not improving. One of the four participants (25%) died after spending some days in the hospital. Deaths related to sepsis are higher in low- and middle-income countries (17), and global fatalities resulting from sepsis have been estimated at 26% (18). These findings underscore the need to better understand the pathogenesis of the disease from a microbiota perspective.
Microbial Composition and Sepsis
The early period of life has been identified as a crucial phase for microbiota to colonize the gut (19), and studies have shown that the gut microbiota composition is shaped by diet and other factors in infancy (20). Therefore, it is essential to investigate the gut microbiota of children presenting with sepsis in Nigeria to better understand the pathogenesis of the disease and develop effective interventions.
The results of this study revealed a marked alteration in the gut microbiota of children with sepsis, demonstrating that the abundance of certain bacterial phyla and families significantly differs from that of healthy individuals. Notably, there was a reduced presence of Bacteroidetes and an increased abundance of Enterobacteriaceae in sepsis cases.
The gut microbiota of weaned participants (controls) was found to be dominated by certain bacterial taxa compared to those exclusively on breast milk or those on breast milk and other foods. For instance, Participant S2 (2 months old) and S4 (2 weeks old) had a microbiota dominated by Bifidobacterium, while in Participant S8, who was consuming breast milk and other foods, the dominance of Bifidobacterium was diminishing. This aligns with the reported association of breastfeeding with microbiota composition and Bifidobacterium-enriched gut during infancy (19).
Health Implications
Dysbiosis of the gut microbiota has been implicated in sepsis (18), characterized by a relative change or alteration in the composition of the gut microbiota with the depletion of beneficial bacterial taxa and an increased population of pathogenic taxa (21). In this study, Megasphaera spp., associated with bacterial vaginosis and pregnancy complications, was dominant in the microbiota of S2 and S8 with sepsis (22). Similarly, Sutterella spp., implicated in degrading immunoglobulin A in ulcerative colitis (23), was found to be dominant in the microbiota of participant S4, also with sepsis. Furthermore, Clostridioides difficile was another pathogenic bacterium that dominated the microbiota of Participant S4. The observed reduction in beneficial bacterial species in participants with sepsis correlates with previous reports of gut microbiota disturbance, with a higher presence of opportunistic pathogens in children with sepsis (24).
Understanding these microbial imbalances is essential for comprehending the underlying mechanisms of sepsis in young children and developing effective interventions. The role of gut dysbiosis in sepsis and its potential as an auxiliary prognostic marker for sepsis in children have been the focus of recent research, emphasizing the significance of evaluating the gut microbiota in the context of sepsis (25).
Global Public Health Relevance
The results of this study highlight the potential health implications of such microbial shifts. A decrease in beneficial commensals, coupled with an increase in pathogenic bacterial species, indicates a concerning pattern in the gut microbiota. These microbial changes could contribute to the severity and progression of sepsis, underscoring the need for early detection and intervention.
The study’s emphasis on the situation in Lagos State, Nigeria, is important. Sepsis is a global health issue that affects vulnerable populations differently in various regions. In Nigeria, sepsis is particularly challenging due to limited healthcare resources, and this study sheds light on the specific dynamics of the disease in this context.
Limitations and Future Research
It is essential to acknowledge the limitations of this study, including its small sample size. Future research should aim to expand the sample to further explore the microbial aspects of sepsis in young children in Nigeria and other resource-limited settings.
Conclusion
The results of this study confirm alterations in the gut microbiota of children presenting with sepsis, characterized by a reduction in beneficial commensals and an increase in potential pathogenic bacterial species.
Our findings have illuminated the significance of understanding the microbiota within the gastrointestinal tracts of young sepsis patients. By examining the microbial communities and structures present in these vulnerable children, we have taken a crucial step toward uncovering the potential links between the composition of gut bacteria and the onset of sepsis. This knowledge has the potential to inform future healthcare strategies, interventions, and treatments for sepsis in young children in Nigeria, particularly in regions with limited healthcare resources.
The report on these four cases highlights the need for further research and underscores the importance of tailored approaches to managing sepsis in this age group. By deepening our understanding of the microbial aspects of this condition, we may ultimately improve the outcomes and quality of care for young sepsis patients in Lagos State and beyond.
Authors’ Contribution
Conceptualization: Abraham Ajayi, Tenny Obiageli Gladys Egwuatu, Stella Ifeanyi Smith.
Data curation: Abraham Ajayi, Uzoma Nonyelum Okeke.
Formal analysis: Abraham Ajayi, Utibeima Udo Essiet.
Investigation: Uzoma Nonyelum Okeke, Sakinat Bello, Awosipe Ayobola Oluwaseun, Abraham Ajayi, Nnenna Kalu.
Methodology: Uzoma Nonyelum Okeke, Sakinat Bello, Awosipe Ayobola Oluwaseun, Abraham Ajayi, Utibeima Udo Essiet, Agatha David, Nnenna Kalu.
Project administration: Abraham Ajayi.
Supervision: Tenny Obiageli Gladys Egwuatu, Stella Ifeanyi Smith, Afolabi Lesi.
Writing–original draft: Abraham Ajayi, Uzoma Nonyelum Okeke.
Writing–review & editing: Tenny Obiageli Gladys Egwuatu, Afolabi Lesi, Stella Ifeanyi Smith.
Competing Interests
The authors declare that there is no conflict of interests.
Data Availability Statement
The raw Illumina sequence reads generated in this study have been deposited in the sequence read archive database of NCBI under BioProject with accession number PRJNA1000308.
Ethical Approval
Approval for this study was obtained from the Institutional Review Board (IRB) of the Nigerian Institute of Medical Research (NIMR) with approval number IRB/21/077 and the Health Research Ethics Committee of the FMC Ebute-Metta with approval number HREC 22-03.
Funding
No funding was obtained for this study.
References
- Sommer MO. Advancing gut microbiome research using cultivation. Curr Opin Microbiol 2015; 27:127-32. doi: 10.1016/j.mib.2015.08.004 [Crossref] [ Google Scholar]
- Smirnov KS, Maier TV, Walker A, Heinzmann SS, Forcisi S, Martinez I. Challenges of metabolomics in human gut microbiota research. Int J Med Microbiol 2016; 306(5):266-79. doi: 10.1016/j.ijmm.2016.03.006 [Crossref] [ Google Scholar]
- Terekhov SS, Smirnov IV, Malakhova MV, Samoilov AE, Manolov AI, Nazarov AS. Ultrahigh-throughput functional profiling of microbiota communities. Proc Natl Acad Sci U S A 2018; 115(38):9551-6. doi: 10.1073/pnas.1811250115 [Crossref] [ Google Scholar]
- Bailey MJ, Naik NN, Wild LE, Patterson WB, Alderete TL. Exposure to air pollutants and the gut microbiota: a potential link between exposure, obesity, and type 2 diabetes. Gut Microbes 2020; 11(5):1188-202. doi: 10.1080/19490976.2020.1749754 [Crossref] [ Google Scholar]
- Zhuang ZQ, Shen LL, Li WW, Fu X, Zeng F, Gui L. Gut microbiota is altered in patients with Alzheimer’s disease. J Alzheimers Dis 2018; 63(4):1337-46. doi: 10.3233/jad-180176 [Crossref] [ Google Scholar]
- Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 2017; 5(1):10. doi: 10.1186/s40168-016-0225-7 [Crossref] [ Google Scholar]
- Meng J, Banerjee S, Li D, Sindberg GM, Wang F, Ma J. Opioid exacerbation of gram-positive sepsis, induced by gut microbial modulation, is rescued by IL-17A neutralization. Sci Rep 2015; 5:10918. doi: 10.1038/srep10918 [Crossref] [ Google Scholar]
- Kullberg RFJ, Wiersinga WJ, Haak BW. Gut microbiota and sepsis: from pathogenesis to novel treatments. Curr Opin Gastroenterol 2021; 37(6):578-85. doi: 10.1097/mog.0000000000000781 [Crossref] [ Google Scholar]
- World Health Organization (WHO). Global Report on the Epidemiology and Burden of Sepsis: Current Evidence, Identifying Gaps and Future Directions. Geneva: WHO; 2020.
- Otu A, Elston J, Nsutebu E. Sepsis in Africa: practical steps to stem the tide. Pan Afr Med J 2015; 21:323. doi: 10.11604/pamj.2015.21.323.6462 [Crossref] [ Google Scholar]
- Nwankwor OC, McKelvie B, Frizzola M, Hunter K, Kabara HS, Oduwole A. A national survey of resources to address sepsis in children in tertiary care centers in Nigeria. Front Pediatr 2019; 7:234. doi: 10.3389/fped.2019.00234 [Crossref] [ Google Scholar]
- Ogundare E, Akintayo A, Aladekomo T, Adeyemi L, Ogunlesi T, Oyelami O. Presentation and outcomes of early and late onset neonatal sepsis in a Nigerian hospital. Afr Health Sci 2019; 19(3):2390-9. doi: 10.4314/ahs.v19i3.12 [Crossref] [ Google Scholar]
- Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol 2017; 2(2):135-43. doi: 10.1016/s2468-1253(16)30119-4 [Crossref] [ Google Scholar]
- Oyetibo GO, Ige OO, Obinani PK, Amund OO. Ecological risk potentials of petroleum hydrocarbons and heavy metals shape the bacterial communities of marine hydrosphere at Atlantic Ocean, Atlas Cove, Nigeria. J Environ Manage 2021; 289:112563. doi: 10.1016/j.jenvman.2021.112563 [Crossref] [ Google Scholar]
- Obi CC, Adebusoye SA, Ugoji EO, Ilori MO, Amund OO, Hickey WJ. Microbial communities in sediments of Lagos Lagoon, Nigeria: elucidation of community structure and potential impacts of contamination by municipal and industrial wastes. Front Microbiol 2016; 7:1213. doi: 10.3389/fmicb.2016.01213 [Crossref] [ Google Scholar]
- Buraimoh OM, Adewumi GA, Akinyemi NM, Amund OO, Ilori MO, Michel FC Jr. Analysis of decomposing wood wastes in the Lagos Lagoon using terminal-restriction fragment length polymorphism and illumina sequencing platform. Niger J Technol Dev 2022; 19(2):116-27. doi: 10.4314/njtd.v19i2.3 [Crossref] [ Google Scholar]
- Carvalho MJ, Sands K, Thomson K, Portal E, Mathias J, Milton R. Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Nat Microbiol 2022; 7(9):1337-47. doi: 10.1038/s41564-022-01184-y [Crossref] [ Google Scholar]
- Sun T, Wang L, Zhang H. Intestinal microbiota in sepsis. Intensive Care Res 2022; 2(1):1-7. doi: 10.1007/s44231-022-00001-8 [Crossref] [ Google Scholar]
- Ou Y, Belzer C, Smidt H, de Weerth C. Development of the gut microbiota in healthy children in the first ten years of life: associations with internalizing and externalizing behavior. Gut Microbes 2022; 14(1):2038853. doi: 10.1080/19490976.2022.2038853 [Crossref] [ Google Scholar]
- Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 2016; 11(6):e0158498. doi: 10.1371/journal.pone.0158498 [Crossref] [ Google Scholar]
- Bassetti M, Bandera A, Gori A. Therapeutic potential of the gut microbiota in the management of sepsis. Crit Care 2020; 24(1):105. doi: 10.1186/s13054-020-2780-3 [Crossref] [ Google Scholar]
- Glascock AL, Jimenez NR, Boundy S, Koparde VN, Brooks JP, Edwards DJ. Unique roles of vaginal Megasphaera phylotypes in reproductive health. Microb Genom 2021; 7(12):000526. doi: 10.1099/mgen.0.000526 [Crossref] [ Google Scholar]
- Kaakoush NO. Sutterella species, IgA-degrading bacteria in ulcerative colitis. Trends Microbiol 2020; 28(7):519-22. doi: 10.1016/j.tim.2020.02.018 [Crossref] [ Google Scholar]
- Du B, Shen N, Tao Y, Sun S, Zhang F, Ren H. Analysis of gut microbiota alteration and application as an auxiliary prognostic marker for sepsis in children: a pilot study. Transl Pediatr 2021; 10(6):1647-57. doi: 10.21037/tp-21-51 [Crossref] [ Google Scholar]
- Weiss SL, Bittinger K, Lee JJ, Friedman ES, Mattei LM, Graham K. Decreased intestinal microbiome diversity in pediatric sepsis: a conceptual framework for intestinal dysbiosis to influence immunometabolic function. Crit Care Explor 2021; 3(3):e0360. doi: 10.1097/cce.0000000000000360 [Crossref] [ Google Scholar]