Avicenna Journal of Clinical Microbiology and Infection. 9(3):97-102.
doi: 10.34172/ajcmi.2022.3390
Original Article
Prevalence and Molecular Mapping of Multidrug-resistant Mycobacterium tuberculosis in Sokoto, North-Western Nigeria
Shuaibu Abdullahi Hudu 1, *  , Abdulgafar Olayiwola Jimoh 2, Yahaya Mohammed 1
, Abdulgafar Olayiwola Jimoh 2, Yahaya Mohammed 1
Author information:
1Department of Medical Microbiology and Parasitology, Faculty of Basic Clinical Sciences, College of Health Sciences, Usmanu Danfodiyo University Sokoto, 840232 Sokoto State, Nigeria
2Department of Pharmacology and Therapeutics, Faculty of Basic Clinical Sciences, College of Health Sciences, Usmanu Danfodiyo University, Sokoto, Nigeria
*
Corresponding author: Shuaibu Abdullahi Hudu, Department of Medical Microbiology and Parasitology, Faculty of Basic Clinical Sciences, College of Health Sciences, Usmanu Danfodiyo University Sokoto, 840232 Sokoto State, Nigeria. Phone: +2348039099312, Email:
hudu.shuaibu@udusok.edu.ng
Abstract
Background: The emergence of multidrug-resistant tuberculosis tuberculosis (MDR-TB) poses a significant danger to Nigeria’s TB control efforts. Nigeria records 570000 new TB cases each year, and it is one of the world’s greatest TB-burden countries as well as one of the top ten countries with MDR-TB. This study aimed to determine the MDR prevalence, phylogenetic analysis, and molecular mapping as well as the link between MDR prevalence and demographic data.
Methods: The study comprised 100 TB patients recruited consecutively. The proportion method on Lowenstein-Jensen (LJ) medium was used for drug susceptibility testing. Conventional polymerase chain reaction (PCR) was used, and the rrs genes were amplified and sequenced. Multiple sequence alignment techniques were used to compare the PCR product sequences to reference sequences retrieved from GenBank.
Results: Rifampicin (RIF) resistance was found in 29% (22.75), ethambutol resistance was found in 20% (15.75), and isoniazid (INH) resistance was found in 28% (21.75). Further, RIF and INH resistance were found in 20% (15.75) of the samples. At nucleotide position 892, four isolates (31%) contained a G-A transition, and the most prevalent mutation found in the rrs gene was S531L (80%, 12.15). The phylogenetic analysis indicated that three of the Sokoto isolates are closely linked to reference isolates from Iran, Germany, China, and Sudan in terms of geographical relatedness.
Conclusions: The obtained data revealed that acquired resistance is a major factor in the establishment of MDR-TB in Sokoto, which could be owing to poor adherence to medication or poor treatment of TB patients.
Keywords: Resistance, Tuberculosis, Multi-drug, Sokoto
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Hudu SA, Jimoh AO, Mohammed Y. Prevalence and molecular mapping of multidrug-resistant Mycobacterium tuberculosis in sokoto, north-western nigeria. Avicenna J Clin Microbiol Infect. 2022; 9(3):97-102. doi:10.34172/ajcmi.2022.3390
Introduction
Multidrug-resistant tuberculosis (MDR-TB) is a global public health problem that jeopardized years of progress in TB control (1). The World Health Organization (WHO) predicted that 18% and 3.5% of treatment experiences and treatment naïve TB patients are MDR, resulting in 558 000 new cases and 230 000 deaths (2). Nigeria is ranked seventh among 30 countries with the greatest TB burden and second in Africa (3). According to the global TB report, an estimated 440 000 new cases of TB were reported in Nigeria in 2019 (of whom 46 000 were HIV-positive), and over 150 000 Nigerians died from the disease (4). In Nigeria, TB is responsible for more than 10% of all deaths. Despite the availability of modern medicines, about 30 individuals die from the condition every hour (5). According to the Federal Ministry of Health, Sokoto State had the highest estimated prevalence of TB in Nigeria in 2016 with 127 cases per 100 000 people. According to the research, Sokoto metropolis has a greater rate of TB cases compared to the rest of the state’s local governments (6).
Resistance to rifampicin (RIF) and isoniazid is known as MDR-TB (7). RIF is a broad-spectrum antibiotic that is still the most effective treatment for Mycobacterium tuberculosis. RIF resistance is caused by a mutation in the rpoB gene, which codes for the RNA polymerase -subunit (8). Isoniazid (INH) has the strongest bactericidal action against TB, is well tolerated, and is inexpensive (9). It is a prodrug that needs to be activated by a catalase-peroxidase enzyme expressed by the KatG gene in mycobacteria (10). Nicotinamide adenine dinucleotide hydrogen dependent enoyl acyl carrier protein reductase encoded by inhA promoter has been inhibited by activated INH inhibiting critical mycoloic. Similarly, INH has been also found to be associated with mutations in katG and inhA gene promoters (11-13). Accordingly, this study aimed to determine the prevalence, molecular characterization, and mapping of MDR-TB.
Methods
This cross-sectional study was conducted from May 2020 to February 2021. Sputum samples were collected prospectively from hundred treatment-naive patients.
Study Area
Sokoto is a major city located in far north-western Nigeria near the confluence of the Sokoto and Rima rivers. It had a population of over 427 760 people in 2006. Sokoto is the current capital of Sokoto State and was formerly the capital of the north-western states. The climate in Sokoto is hot and semi-arid. It is flanked by sand savannah and lonely hills in the parched Sahel. Sokoto is one of the warmest cities in Nigeria with an annual average temperature of 28.3°C. However, maximum daytime temperatures are normally below 40°C (104.0°F) for the majority of the year, and the dryness makes the heat bearable. February to April are the hottest months with daytime temperatures reaching 40°C. The maximum temperature ever recorded was 45°C. The rainy season lasts from June to October, and rainfall is almost every day. The showers do not linger long and are not like the constant heavy rainfall that may be found in many tropical areas. During the cold season, from late October to February, the harmattan wind blows Sahara dust across the region, dictating the climate. The dust reduces the amount of light that reaches the ground and decreases temperatures dramatically. According to a 2006 census, the population of Sokoto state is estimated to be 3.7 million people and is divided into two ethnic groups: Fulani and Hausa. Sokoto town, the state capital, has a population of about 2.5 million people. In the local government border areas, there are Zabarmawa and Tuareg populations in addition to Fulani and Hausa. The Hausa language is spoken by all of these ethnicities, but the Fulani people speak Fulfulde. Gobirawa, Zamfarawa, Kabawa, Adarawa, and Arawa are Hausa tribes in the state. The Fulani, on the other hand, is divided into two groups: town Fulani (Hausa: Fulani Gida; Fula: Fule Wuro) and nomads (Hausa: Fulani Gida; Fula: Fule Wuro). The Torankawa, Shehu Usmanu Danfodiyo’s clan, Sullubawa, and Zoramawa belong to the former. The Tonkawa has been the aristocratic class since 1804.
Mycobacterium tuberculosis Culture and Sensitivity
Approximately 4 to 5 mL of sputum were transferred to McCartney tubes, and double the amount of sterile NaOH solution was added. The McCartney tube caps were then fastened, and the mixture was thoroughly mixed. The bottles were shaken and incubated at 37°C for 15 minutes. After 20 minutes, the McCartney tubes were taken out of the incubator, 15 mL of sterile distilled water was added, and the mixture was thoroughly mixed before the tubes were centrifuged at 3000 × g for 15 minutes. After washing the pellet with sterile water at 3000 × g for 15 minutes and the supernatant was discarded from the sediment, slopes of Lowenstein-Jensen (LJ) medium were infected with the supernatant using a sterile wire loop and incubated at 37°C. According to the WHO laboratory recommendations, sputum cultures and LJ medium microscopy analyses were performed (14).
GeneXpert Assay
It is an automated polymerase chain reaction (PCR) base test that may detect the DNA of mycobacterium and the RIF resistance gene. GeneXpert is utilized in this study because it is an outstanding clinical tool for the early diagnosis of RIF resistance as well as the early detection of M. tuberculosis even when a direct smear is negative. It can determine whether someone has a TB infection and whether their TB bacterium is resistant to one of the most widely used TB medications such as RIF. To conduct this assay, the sputum collection container’s lid was unscrewed and turned upside down to prevent sputum from spilling onto the counter. After adding GeneXpert reagent to the container in an amount equivalent to the amount of sputum inside the container, the sputum collection container’s lid was screwed back on. Then, the mixture was shaken 10-20 times clockwise and set aside at room temperature for 15 minutes. This operation continued until the sputum liquefied to a water-like consistency. Afterwards, the GeneXpert cartridge was positioned upright on the table, the blue lid of the labelled cartridge was opened, and a sterile pipette was used to slowly pipette sputum into the cartridge. The GeneXpert machine was turned on, the cartridge barcode was scanned, the cartridge was put into the machine and allowed to run for 105 minutes, and the final reading was performed.
DNA Extraction
Following the manufacturer’s instructions, DNA was extracted from a sputum sample using the MagMAX Viral RNA Isolation Kit (Applied Biosystems, Ambion, USA) (12,13). Then, NonoDrop was used to evaluate the concentration and purity of the extracts (A260/A280 nm and A260/A230 nm) (Thermo Fisher Scientific, USA).
PCR Amplification of rrs Gene
Genomic DNA was used as a template for PCR amplification of the whole rrs gene. F- (5 -AGAGTTTGATCCTGGCTCAG-3) and R-(5 -CTACGGCTACCTTGTTACGA-3) were used as forward and reverse primers, respectively. The choice of the rrs gene was due to the significance of its mutation in geographic variation, which makes it a gene of interest in phylogenetic analysis. The rrs gene alterations are also linked to streptomycin resistance in TB. As reported in Table 1, the reaction volume was created in 25 L thin wall microtubes containing a PCR reaction mixture. The target gene was amplified using an automated PCR thermocycler with the following PCR conditions: denaturation at 95°C for 3 minutes, followed by 30 PCR cycles of denaturation for 30 seconds at 96°C, annealing for 30 seconds at 60°C, and extension for 1 minute and 30 seconds at 72°C. The PCR products were purified using a commercial kit according to the manufacturer’s guidelines (Vivantis Technologies, USA).
Table 1.
Reaction Mixture Used in the PCR Amplification of HCV
|
Reagent
|
Volume/Reaction
|
Final Concentration
|
| dNTPs mix |
0.5 µL |
0.8 mM |
| Fast Taq polymerase |
0.5 µL |
0.1 U |
| MgSO4 (25mM) |
2.0 µL |
2 mM |
| Forward primer |
0.3 µL |
0.3 µM |
| Reverse primer |
0.3 µL |
0.3 µM |
| Buffer (Tri base, EDTA, and acetic acid) |
5.0 µL |
1x |
| cDNA template |
4 µL |
- |
| RNase free water |
12.4 µL |
- |
| Total |
25 µL |
|
Note. PCR: Polymerase chain reaction; HCV: Hepatitis C virus.
Sequencing of 16S rRNA
The purified fragments and PCR products were sequenced commercially by integrated DNAT using Sanger sequencing methods (IDT, Gemini Singapore). The Sokoto MDRTB isolate sequence was deposited in the GenBank with accession numbers ON025967, ON025968, ON025969, ON025970, ON025971, and ON025972. The partial sequence of rr-gene from the MDRTB positive samples was subjected to nucleotide blast search for sequence homology using the BLASTN. The E-value score and sequence identity were among the parameters evaluated for sequence homologies. Sequences with a lower E-value score (<10-3) and identity >70% were considered biologically homologous (Brenner et al (15)).
Phylogenetic Analysis
Multiple sequence alignment of the local MDRTB isolate and the reference isolates retrieved from GeneBank was performed using the MEGA11 (16). The reliability of the sequence alignment was verified by computing the overall mean distance which is the number of base differences per site from the average overall sequence pair. Nucleotide p-distance < 0.33 was considered the best sequence alignment. A phylogenetic analysis was performed using the neighbour joining and the maximum likelihood methods with the aid of the MEGA11 software (16).
Data Analysis
The data were collected and analyzed using the statistical program IMB SPSS. Descriptive and inferential statistics were used in the form of frequencies and percentages to characterize the data. The chi-square test was used to examine the statistical significance of the TB recovery rate among males and females, and P value of 0.05 was considered statistically significant.
Results
Twenty revealed no growth, but 2 sputum samples were developed in p-nitro benzoic acid, suggesting the existence of a Mycobacterium not related to TB. The rate of growth on LJ medium was as follows: 39% of respondents (29.75) had +3 colonies (200–500), 33% of the respondents (25.75) had +2 colonies (100–200), 26.7% (20.75) had +1 colony (10–99), and 1.3% (1.75) had 4 colonies. The preponderance of MDR growth seemed to be positive (value 0.426), demonstrating a statistically insignificant link between the rate of growth and MDR.
Moreover, MDR was discovered in 20% (15.75) of selected patients and old patients, respectively, and 14.7% (11.15) of new cases were diagnosed (value 0.003), demonstrating that drug resistance is associated with previous treatment. Further, MDR cases were found in 73.3% of men and 26.7% of women (value 0.536). In addition, 60% of MDR cases (9.15) were found in people under the age of 30 (value 0.652) as illustrated in Figure 1.
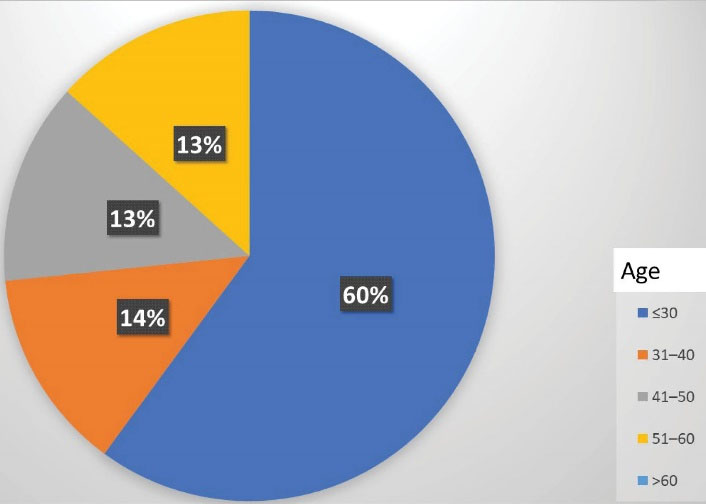
Figure 1.
Distribution of MDR Among Age Groups. Note. MDR: Multi-drug resistance
.
Distribution of MDR Among Age Groups. Note. MDR: Multi-drug resistance
Streptomycin resistance was found in 56% (42.75) of M. tuberculosis isolates, and RIF resistance was found in 29% (22.75). Likewise, ethambutol resistance was found in 20% (15.75), and INH resistance was found in 28% (21.75) of M. tuberculosis isolates. Further, RIF and INH resistance were found in 20% (15.75) of the samples. Only 16% (12.75) of the patients were resistant to all four medications. Under a UV transilluminator documentation machine, the run evolution of the final separation of PCR products was visualized. The majority of them fluoresced brightly, while 2 fluoresced slightly and were eliminated.
The most common mutation was S531L (80%, 12.15) followed by S315T (67%, 10.15), and it seemed that 62% of streptomycin-resistant isolates (8.13) reveal rrs gene alterations, which were divided into 7 groups. As presented in Table 2, three isolates (23%) have a C-A transversion at nucleic acid position 222, 4 isolates (31%) have a G-A transformation at nucleic acid position 892, 1 isolate (8%) has a G-T transversion at nucleic acid position 855. Additionally, one isolate (8%) has an A-G transformation at nucleotide position 906, one isolate (8%) has a T-A transformation, and two isolates have G-A. Moreover, phylogenetic analysis demonstrates the homology of the local sequences in terms of geographical relatedness to isolates from other parts of the world as illustrated in Figure 2. Further, Figure 3 depicts mapping of the Sokoto isolate according to local government.
Table 2.
The Resistance Pattern of Isolates Explaining the Location and Frequency of Polymorphisms
|
Nucleotide Mutation and Position
|
The Proportion of the Mutation (%)
|
| C222A |
23 |
| G885T |
8 |
| G892A |
31 |
| A904G |
15 |
| A906G |
8 |
| T906A |
8 |
| G1400A |
8 |
| G 1400C |
8 |
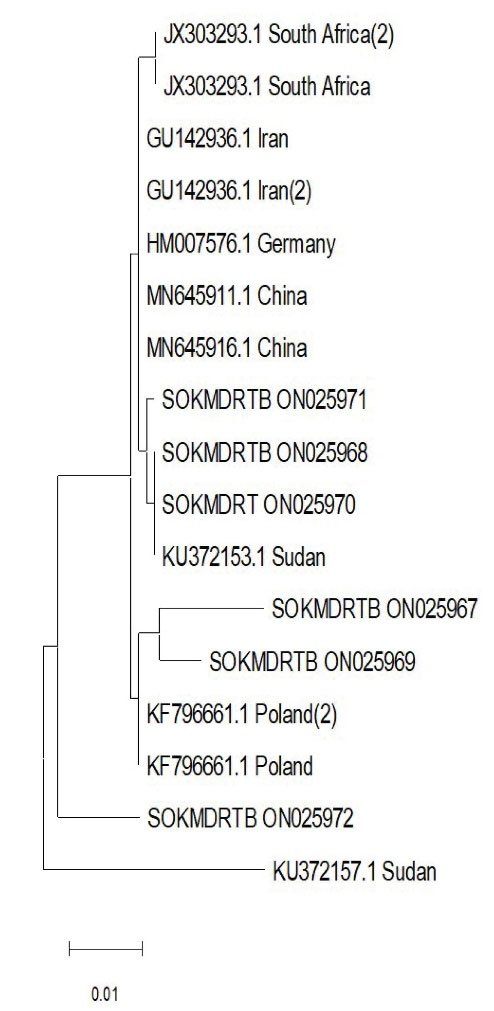
Figure 2.
Phylogenetic tree of Local Isolate with Reference Isolates. The Topologies with The Maximum Log-Likelihood Value Were Chosen After the Neighbour-Join and Bionj Algorithms Were Performed to A Matrix of Pairwise Distances Estimated Using the Tamura-Nei Model.
.
Phylogenetic tree of Local Isolate with Reference Isolates. The Topologies with The Maximum Log-Likelihood Value Were Chosen After the Neighbour-Join and Bionj Algorithms Were Performed to A Matrix of Pairwise Distances Estimated Using the Tamura-Nei Model.
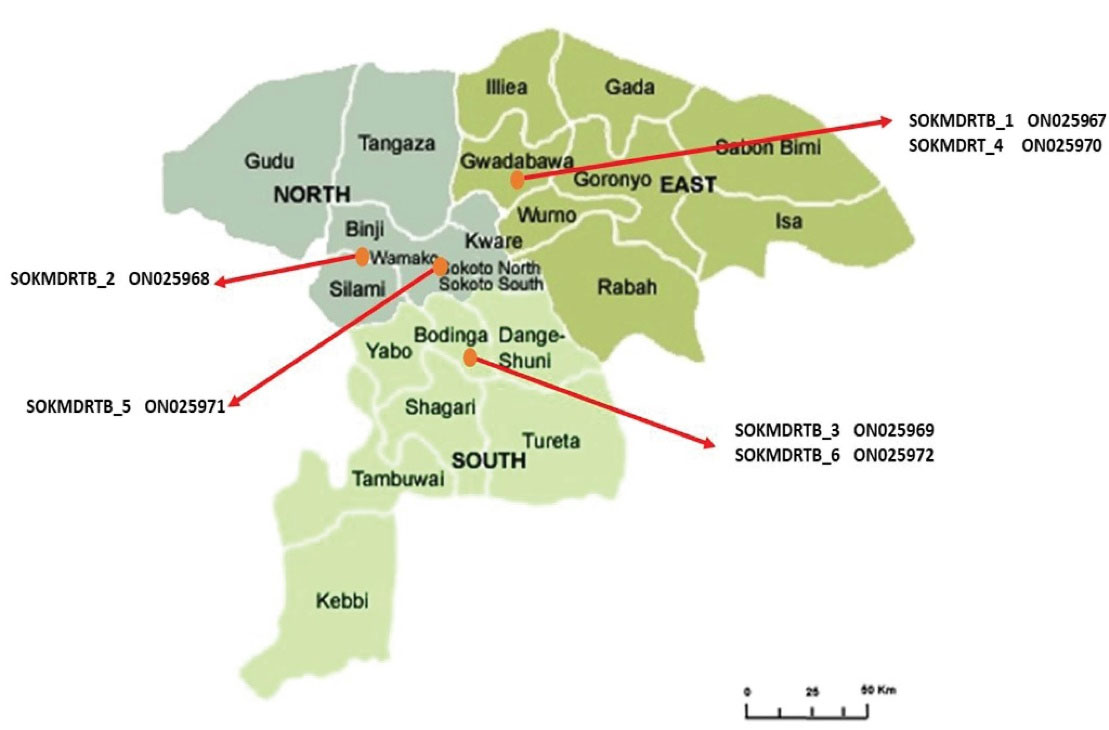
Figure 3.
Map of Sokoto State Showing Local Government of MDR Isolate. Note. MDR: Multi-drug Resistance
.
Map of Sokoto State Showing Local Government of MDR Isolate. Note. MDR: Multi-drug Resistance
Discussion
A novel C/A transversion mutation was identified at position 222 throughout the study. This may be the first study to demonstrate 16S rRNA evaluation among MDR M. tuberculosis strains isolated from Nigeria, and the prevalence of MDR was 20% (15.75) and 14.7% (11.15) in old cases, respectively, and 5.3% (4.15) in new diagnosed cases, which was lower than a study in France that reported a prevalence of 7.1% (23.323). Re-treated patients accounted for 31.4% (33.105) of the total (17), and this difference could be related to geographic differences. Further, males accounted for 73.3% of MDR patients in this study, while females accounted for 26.7%, which is consistent with Raizada and colleagues’ finding in which MDR-TB was more prevalent (72%, 230.320) in males (18). However, this result is in contrast to Melzer and colleagues’ study which found 56.7% and 43.3% prevalence in males and females, respectively (19). This could be due to a large number of males in this study compared to females. Moreover, the majority of MDR cases (60%) were discovered in people under the age of 30 as reported previously (19,20).
In addition, a total of 23% of the isolates were found to have mutations in the 912 areas of the rrs gene as the most common mutation in this region of Africa. The rrs genetic variant frequency of 62% was also found, which is similar to a study in Sudan which discovered 70% (35.50) strains (21). Moreover, the highest frequency was found in China, Zambia, and Latvia with a prevalence of 85.7%, 77.8%, and 85%, respectively (22-25). This might be associated with international trade travel between China and most of the African countries, and travellers who interacted with people diagnosed with MDR TB were most at risk. However, there was a low risk of contracting any form of TB just by flying. Studies in Germany revealed that a G transition occurs at position 904 (26). However, no mutations in the rrs 912 area were found in studies conducted in Barcelona (27). The studies conducted in New York (20) and Poland (28) both agreed that the mutation in the rrs coding area at loop 912 has not been discovered. The absence of rrs 912 mutations in these countries and its presence in the current study and other African countries suggest a significant geographical difference between African variants and the variants found in Europe. This rrs 912 mutation has the potential to be used as a predictive biomarker, a tool for promptly monitoring streptomycin resistance and a tool for doctors. Since the small sample size is one of the limitations of the present study, higher sample sizes in future research could be useful for identifying other markers that contribute to the diagnosis of streptomycin-resistant MTB.
According to phylogenetic analysis, 3 Sokoto isolates were found to be closely related to reference isolates from Sudan, China, Germany, and Iran. Although they belong to the same clade as an isolate from Poland, SOKMDRTB 1 ON025967 and SOKMDRTB 3 ON025969 exhibit some evolutionary distance in a sub-clade. This is not surprising given the significant number of Sokoto residents who travel to Iran and Sudan to pursue both the Islamic and Western education, opening up a potential route for the transmission of these mutations and explaining the genetic similarity. Similar to the previous strain, isolate SOKMDRTB 6 ON025972 is part of a different clade; however, all of the isolates are probably descended from Sudan isolates with accession number KU372157. The two isolates found in the Bodinga local government area are SOKMDRTB 3 ON025969 (closely linked to the Polish isolate) and SOKMDRTB 6 ON025972 (a unique isolate originating from the Sudan isolate). Two further isolates are SOKMDRTB 1 ON025967 (similarly related to Poland isolate) and SOKMDRT 4 ON025970 (closely connected to Sudan isolate) which can be found in the Gwadabawa local government region. Although the relationship between Poland and the Bodinda and Gwadabawa Isolates is unclear, infectious diseases make no difference either in national or regional boundaries. For example, one isolate found in Wamakko and Sokoto North was closely related to those from China and Sudan.
Conclusions
A serious threat to global TB control efforts is MDR-TB. Further, the information regarding the molecular genetics of the strains recovered in Sokoto is scarce. The present findings indicated that acquired resistance, which may be caused by poor medication adherence or subpar management of TB patients, is a significant contributing factor to the development of MDR-TB in Sokoto in addition to the extraordinary transmission. Rapid identification and adherence to a suitable treatment strategy are necessary for managing MDR-TB in Sokoto. Further, as a result of commercial or academic trips, the majority of the Sokoto isolates were found through phylogenetic analysis to be closely related to reference isolates from Sudan, China, and Iran.
Acknowledgements
The authors wish to acknowledge Tertiary Education Trust Fund, Nigeria, for providing the TET Fund Institutional Base Research Grant for this study with Grant No. TETFUND/DR&D/CE/UNIV/SOKOTO/RP/VOL.1
Conflict of Interests
None to be declared.
Ethical Approval
Ethical approval was obtained from the Specialist Ethical committee, Sokoto (Ref. SHS/SUB/133/VOL1).
References
- Espinosa-Pereiro J, Sánchez-Montalvá A, Aznar ML, Espiau M. MDR Tuberculosis Treatment. Medicina 2022; 58(2):188. doi: 10.3390/medicina58020188 [Crossref] [ Google Scholar]
- WHO. Meeting report of the WHO expert consultation on the definition of extensively drug-resistant tuberculosis, 27-29 October 2020. WHO; 2021.
- Angell B, Sanuade O, Adetifa IM, Okeke IN, Adamu AL, Aliyu MH. Population health outcomes in Nigeria compared with other west African countries, 1998-2019: a systematic analysis for the Global Burden of Disease Study. Lancet 2022; 399(10330):1117-29. doi: 10.1016/S0140-6736(21)02722-7 [Crossref] [ Google Scholar]
- Chakaya J, Khan M, Ntoumi F, Aklillu E, Fatima R, Mwaba P. Global Tuberculosis Report 2020–Reflections on the Global TB burden, treatment and prevention efforts. Int J Infect Dis 2021; 113:S7-S12. doi: 10.1016/j.ijid.2021.02.107 [Crossref] [ Google Scholar]
- Ogbuabor DC, Onwujekwe OE. Governance of tuberculosis control programme in Nigeria. Infect Dis Poverty 2019; 8(1):1-11. doi: 10.1186/s40249-019-0556-2 [Crossref] [ Google Scholar]
- Sakajiki MA, Garba B, Ibrahim Y, Mohammed BA, Abdullahi U, Sada KB. Treatment outcome of Tuberculosis at a specialist hospital in North-Western Nigeria-A 30 months retrospective study. Pak J Chest Med 2018; 24(1):04-9. [ Google Scholar]
- Salaam-Dreyer Z, Streicher EM, Sirgel FA, Menardo F, Borrell S, Reinhard M. Rifampicin-monoresistant tuberculosis is not the same as multidrug-resistant tuberculosis: a descriptive study from Khayelitsha, South Africa. Antimicrob Agents Chemother 2021; 65(11):e00364-21. doi: 10.1128/AAC.00364-21 [Crossref] [ Google Scholar]
- Ardizzoni E, Ariza E, Mulengwa D, Mpala Q, de La Tour R, Maphalala G. Thin-Layer-Agar-Based Direct Phenotypic Drug Susceptibility Testing on Sputum in Eswatini Rapidly Detects Mycobacterium tuberculosis Growth and Rifampicin Resistance Otherwise Missed by WHO-Endorsed Diagnostic Tests. Antimicrob Agents Chemother 2021; 65(6):e02263-20. doi: 10.1128/AAC.02263-20 [Crossref] [ Google Scholar]
- Gausi K, Ignatius EH, Sun X, Kim S, Moran L, Wiesner L. A semi mechanistic model of the bactericidal activity of high-dose isoniazid against multidrug-resistant tuberculosis: Results from a randomized clinical trial. Am J Respir Crit Care Med 2021; 204(11):1327-35. doi: 10.1164/rccm.202103-0534OC [Crossref] [ Google Scholar]
- Lang J, Vincent L, Chenel M, Ogungbenro K, Galetin A. Reduced physiologically-based pharmacokinetic model of dabigatran etexilate-dabigatran and its application for prediction of intestinal P-gp-mediated drug-drug interactions. Eur J Pharm Sci 2021; 165:105932. doi: 10.1016/j.ejps.2021.105932 [Crossref] [ Google Scholar]
- Norouzi F, Moghim S, Farzaneh S, Fazeli H, Salehi M, Nasr Esfahani B. Significance of the coexistence of non-codon 315 katG, inhA, and oxyR-ahpC intergenic gene mutations among isoniazid-resistant and multidrug-resistant isolates of Mycobacterium tuberculosis: a report of novel mutations. Pathog Glob Health 2022; 116(1):22-9. doi: 10.1080/20477724.2021.1928870 [Crossref] [ Google Scholar]
- Telenti A, Marchesi F, Balz M, Bally F, Böttger EC, Bodmer T. Rapid identification of mycobacteria to the species level by a polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol 1993; 31(2):175-8. doi: 10.1128/jcm.31.2.175-178.1993 [Crossref] [ Google Scholar]
- Narvaiz de Kantor I, Kim SJ, Frieden TR, Laszlo A, Luelmo F, Norval PY, et al. Laboratory services in tuberculosis control. World Health Organization; 1998.
- Hagemann P. World Health Organization. Manual of Basic Techniques for a Health Laboratory. Clin Chem 2003;49:1712-3. Available from: https://www.degruyter.com/document/doi/10.1515/labm.2004.28.1.101/html.
- Brenner SE, Chothia C, Hubbard TJ. Assessing sequence comparison methods with reliable structurally identified distant evolutionary relationships. Proceedings of the National Academy of Sciences 1998 May 26; 95(11):6073-8. [ Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021; 38(7):3022-7. doi: 10.1093/molbev/msab120 [Crossref] [ Google Scholar]
- Lyu J, Kim M, Song J, Choi C, Oh Y, Lee S. GenoType® MTBDRplus assay detection of drug-resistant tuberculosis in routine practice in Korea. Int J Tuberc Lung Dis 2013; 17(1):120-4. doi: 10.5588/ijtld.12.0197 [Crossref] [ Google Scholar]
- Raizada N, Sachdeva K, Chauhan D, Malhotra B, Reddy K, Dave P. A multi-site validation in India of the line probe assay for the rapid diagnosis of multi-drug resistant tuberculosis directly from sputum specimens. PLoS One 2014; 9(2):e88626. doi: 10.1371/journal.pone.0088626 [Crossref] [ Google Scholar]
- Melzer M, Gupta N, Petersen I, Cook S, Hall B. Previous treatment in predicting drug-resistant tuberculosis in an area bordering East London, UK. Int J Infect Dis 2010; 14(8):e717-e22. doi: 10.1016/j.ijid.2010.02.2247 [Crossref] [ Google Scholar]
- Barnard M, Albert H, Coetzee G, O’Brien R, Bosman ME. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am J Respir Crit Care Med 2008; 177(7):787-92. doi: 10.1164/rccm.200709-1436OC [Crossref] [ Google Scholar]
- Sabeel S, Salih M, Ali M, El-Zaki S, Abuzeid N, Elgadi Z. Phenotypic and genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Sudanese patients. Tuberc Res Treat 2017; 2017:8340746. doi: 10.1155/2017/8340746 [Crossref] [ Google Scholar]
- Yang J, Zhang T, Xian X, Li Y, Wang R, Wang P. Molecular characteristics and drug resistance of Mycobacterium tuberculosis isolate circulating in Shaanxi province, Northwestern China. Microb Drug Resist 2021; 27(9):1207-17. doi: 10.1089/mdr.2020.0496 [Crossref] [ Google Scholar]
- Bwalya P, Yamaguchi T, Solo ES, Chizimu JY, Mbulo G, Nakajima C. Characterization of mutations associated with streptomycin resistance in multidrug-resistant Mycobacterium tuberculosis in Zambia. Antibiotics 2021; 10(10):1169. doi: 10.3390/antibiotics10101169 [Crossref] [ Google Scholar]
- Tracevska T, Jansone I, Nodieva A, Marga O, Skenders G, Baumanis V. Characterisation of rpsL, rrs and embB mutations associated with streptomycin and ethambutol resistance in Mycobacterium tuberculosis. Res Microbiol 2004; 155(10):830-4. doi: 10.1016/j.resmic.2004.06.007 [Crossref] [ Google Scholar]
- Singh PK, Singh U, Jain A. Emergence of Specific gyr A Mutations Associated High-Level Fluoroquinolone-Resistant Mycobacterium tuberculosis among Multidrug-Resistant Tuberculosis Cases in North India. Microb Drug Resist 2021; 27(5):647-51. doi: 10.1089/mdr.2020.0240 [Crossref] [ Google Scholar]
- Meier A, Kirschner P, Bange F-C, Vogel U, Böttger E. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother 1994; 38(2):228-33. doi: 10.1128/AAC.38.2.228 [Crossref] [ Google Scholar]
- Tudó G, Rey E, Borrell S, Alcaide F, Codina G, Coll P. Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis clinical isolates in the area of Barcelona. J Antimicrob Chemother 2010; 65(11):2341-6. doi: 10.1093/jac/dkq322 [Crossref] [ Google Scholar]
- Jagielski T, Ignatowska H, Bakuła Z, Dziewit Ł, Napiórkowska A, Augustynowicz-Kopeć E. Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. Plos One 2014; 9(6):e100078-e. doi: 10.1371/journal.pone.0100078 [Crossref] [ Google Scholar]