Avicenna Journal of Clinical Microbiology and Infection. 9(2):88-96.
doi: 10.34172/ajcmi.2022.14
Review Article
A Short Review of Forensic Microbiology
Fatemeh Yousefsaber 1, *  , Zeinab Naseri 2, Amir Hosein Hasani 1
, Zeinab Naseri 2, Amir Hosein Hasani 1
Author information:
1Department of Cell and Molecular Biology and Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran
2Department of Biotechnology, Faculty of Agriculture, Bu-Ali Sina University, Hamedan, Iran
*
Corresponding author: Fatemeh Yousefsaber, Department of Cell and Molecular Biology and Microbiology, Faculty of Biological Science and Technology, University of Isfahan, Isfahan, Iran. Email:
Fatemeh.ysaber@gmail.com
Abstract
Background: Microbial forensics is a multidisciplinary area, which has been recently considered an effective tool in forensic investigations. This growing field of forensics covers a wide spectrum of different branches of science, including biology, chemistry, physics, geology, mathematics, and computer sciences, leading to a practical approach that can be applied in several areas such as bioterrorist actions, environmental issues, emerging and reemerging diseases, as well as reliable trace evidence at a crime scene.
Methods: The information has been gathered via Google Scholar using several keywords, including forensic microbiology, bioterrorism, forensic investigation, and trace evidence. The data were from reliable articles and books published over 50 years. This paper is a short review of forensic microbiology with a bioinformatics perspective to use in different fields such as the court.
Results: It is known that using either microorganisms or their toxins is a low-cost potential tool with serious morbidity and mortality rates that can spread all around the world by food or water supplies or even through the air, making them a perfect candidate bioweapon with minimum traceability. Studies have indicated that environmental conditions plus biological and abiotic factors would affect the following analysis and the final validation, which is an essential step in the forensic investigation due to its highly effective role in the court vote. To face different challenges, law enforcement has the infrastructure for attribution and deterrence (e.g., following the exact microbial forensics program) so that it can be used in court. Developing more reproducible, sensitive, and accurate methods, preparing a wide reliable database, and devoting the right amount of budget will help improve the whole forensic procedure in the legal system.
Conclusions: The current paper is a short review of how forensic scientists can use microbial features on a crime scene to clarify and enhance the procedure to solve different criminal cases.
Keywords: Forensic microbiology, Bioterrorism, Forensic investigation, Trace evidence
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Yousefsaber F, Naseri Z, Hasani AH. A short review of forensic microbiology. Avicenna J Clin Microbiol Infect. 2022; 9(2):88-96. doi:10.34172/ajcmi.2022.14
Introduction
Microbial forensics is defined as the multidisciplinary major of science that uses scientific methods to analyze evidence in case of bioterrorist actions, or the inadvertent release of a pathogenic microorganism or toxin for determined ends (1).
This multidisciplinary major covers a wide variety of different sciences, including biology, chemistry, physics, geology, mathematics, and computer sciences (2).
It is a known fact that the use of either microorganisms or their toxin is a low-cost potential tool with serious morbidity and mortality that can spread worldwide by food or water supplies or even through the air, changing them into a perfect candidate bioweapon with minimum traceability.
Bioterrorism is an intentional use of biological agents, including microbes and their toxins mainly to cause terror and affect theeconomic situation (1).
As long as there is a risk of a bioterrorist action, forensic microbiologists should choose detection methods that are not only accurate to determine the exact agent but quick enough to prevent public panic in society, including DNA microarrays or biochips (3).
To encounter different challenges, law enforcement has the infrastructure for attribution and deterrence (e.g., following the exact microbial forensics program) so that it can be employed in the court (Figure 1). A successful microbial forensic investigation has special characteristics as follows:
-
High sensitivity and specificity for detection and identification;
-
Large rational databases;
-
Validated analytical methods;
-
Following quality assurance guidelines to obtain reliable results.
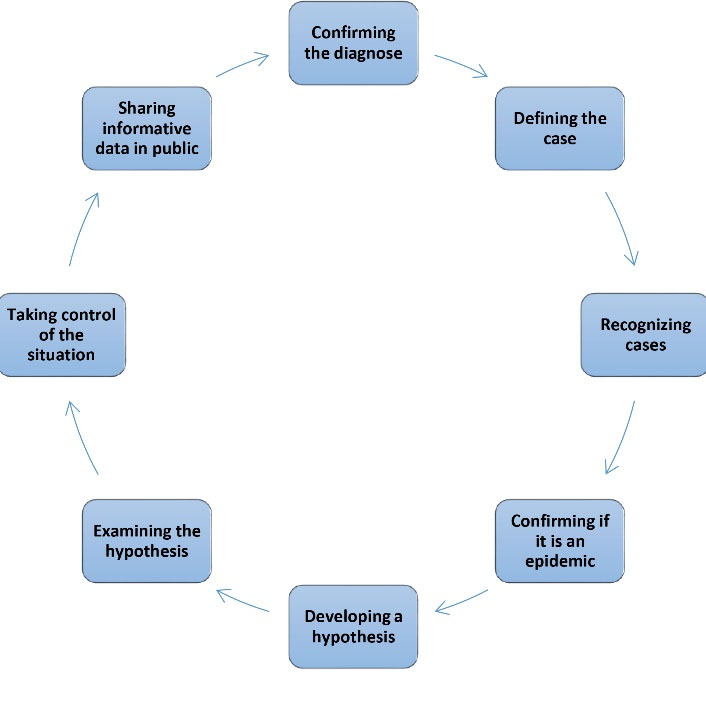
Figure 1.
Recognized Steps for Investigating Contagious diseases. Source. Centers for Disease Control and Prevention (11).
.
Recognized Steps for Investigating Contagious diseases. Source. Centers for Disease Control and Prevention (11).
The present paper is a short review focusing on how forensic scientists can apply microbial features on a crime scene to clarify and enhance the procedure to solve various criminal cases (1).
History
Nearly two thousand years ago, the first bioterrorist activity was recorded, when ancient Romans deliberately poisoned their enemy’s water source by bringing carrion into wells (4).
Although there were several signs of using biological agents in Homer’s epic poems, Iliad and Odyssey, pointing out the fact of tipping arrows and spears with poison.
A few examples of bio crimes in history are the European plague epidemic during the Middle Ages (also known as Black Death), Smallpox and measles against Native Americans, Plague infected rats against the Japanese during World War II, and the International Shigellosis contamination in 1996 in Texas. In addition, the other examples include the anthrax letter in United States, anthrax cases among heroin users in Scotland, Escherichia coli O104:H4 outbreak in Germany, and the Ebola outbreak in 2014 (5-10).
One of the most common medical-related bio crimes involves dentists who infect their patients. In 1992, a group of scientists conducted research on the transmission of the human immunodeficiency virus in a dental practice. According to a phylogenetic analysis of HIV strains isolated from him and his patients, it was clarified that the mentioned dentist was in charge of infecting his patients during oral surgery (3).
Among all instances of microbial attacks in history, the anthrax letter of 2001 draw huge public attention, leading to the foundation of the Scientific Working Group on Microbial Genetics and Forensics on 29 July 2002 hosted by the Federal Bureau of Investigation (FBI) (1).
Applications
As mentioned before, microbial forensics can be applied in various areas from a special medical condition to national security issues (e.g., in a case of a bio-attack). Solving environmental issues, investigating emerging diseases around the world, considering personal identification, determining the cause, time, and location of death, and examining the sexual assault cases are just a few examples of microbial forensics applications, which will be discussed in detail in the following sections.
Transmission of Disease: From Individual to Bioterrorist Action
One of the forensic microbiology applications is perusing the source of disease whether they are endemic, epidemic, or pandemic. The important issue about the natural outbreak or bioterrorist action is the horizontal and vertical transmission, especially the rate of morbidity and mortality which can be divided into direct and indirect transmission categories.
The direct transmission includes direct contact such as touching, biting, sexual intercourse, or even animals and soils in case of zoonosis diseases, and direct projections are sneezing and coughs, leading to droplet spread.
On the other hand, indirect transmission can be listed as blood transfusion, physical or biological spread, and airborne transmission by dust.
To figure out a biological attack, there are some epidemiological clues published in 2003 as follows:
simple
-
- When a disease is caused by an unusual agent;
-
- Emerging uncommon strains of agents;
-
- When a common sickness gets higher morbidity and mortality;
-
- Unusual distribution (either geographically or seasonal);
-
- An uncommon disease affecting a large population, especially for a specified age or group
-
- Similar genetic patterns between isolated agents;
-
- Enormous unexplained illness and death cases (12).
Deliberate Infection of Animals
For over a thousand years, humans have been using animals to serve all sorts of their desires, including food, security, transportation, and the like.
Animals were also involved in deliberate infection; according to ancient history, well poisoning has been documented as the earliest biological warfare. The rotting corpse of horses and rats was thrown down the water source, which was counted as the main reason for plague and tuberculosis transmissions. Nonetheless, some of the historical evidence seems to be remarkably affected by superstitious beliefs (13).
The anthrax letter has been considered the highlight of microbial forensics. Moreover, the role of Germany in the deliberate infection of animals during the world one epic is unavoidable. It has been reported that Dr. Anton Dilger produced a large batch of Bacillus anthracis and Burkholderia mallei agents which later caused infection in more than 3000 horses, cattle, and other animals that were supposed to be sent to European countries, including Romanian, France, and Argentina (14,15).
Rabbit calicivirus disease as a bioagent to reduce the rabbits’ population in New Zealand (1997), the Salmonella Typhimurium contamination of salad bars in Dalles (1984), and emerging a water-born in Canada caused by E. coli O157:H7 and Campylobacter jejuni (2000) are also important cases related to the subject of illegally using animals to introduce a biological agent to specified population or location (16-18). Beside the mentioned cases, there are other microorganisms that had been applied as a bioweapon (Table 1).
Table 1.
Few Identified Bacteria, Viruses, and Toxins That Could Be Used as a Bioweapon
|
Bio Agents
|
Name
|
Disease
|
| Bacteria
|
Bacillus anthracis
|
Anthrax |
|
Clostridium botulinum
|
Botulism |
|
Brucella species
|
Brucellosis |
|
Burkholderia mallei
|
Glanders |
|
Burkholderia pseudomallei
|
Melioidosis |
|
Coxiella burnetii
|
Q fever |
|
Escherichia coli O157:H7 |
Hemolytic uremic syndrome |
|
Francisella tularensis
|
Tularaemia |
|
Salmonella species |
Salmonellosis |
|
Salmonella typhi
|
Typhoid fever |
|
Shigella species |
Shigellosis |
|
Vibrio cholera
|
Cholera |
|
Yersinia pestis
|
Plague |
| Viruses
|
Arenaviruses
|
Junin and Lassa fever |
|
Ebola virus |
Ebola virus haemorrhagic fever |
|
Lassa virus |
Lassa fever |
|
Marburg virus |
Marburg virus haemorrhagic fever |
|
Variola major |
Smallpox |
| Toxins
|
Botulinum toxin |
Botulism |
| Ricin toxin from Ricinus communis |
No explanation |
Source. Hawksworth and Wiltshire (19).
Trace Evidence
There are enormous applications of different branches of microbiology, especially mycology in forensic sciences, including considering post-mortem intervals, determining the cause of death, locating corps, and the like.
One of the most valuable documents, using fungi and their spores, as well as bacterial spores, is tracing evidence, which has been proved to be useful in court (20).
Microbial Decomposition
During the decomposition process, microbial communities will change and affect not only the corpse but also its surrounding environment. It should be noticed that any changes during the time would remain as trace evidence and could be different in each ecosystem.
Studies demonstrated that there is a microbial decomposer pattern, including bacteria, fungi, and non-fungal eukaryotes during decomposition (Figure 2). When the corpse is still fresh, the Proteobacteria is the dominant species but as time goes on, Bacteroidetes and Firmicutes will take the place, as well as Ascomycota fungi (21).

Figure 2.
Microbial Decomposer Clock. Source. Allwood et al (22).
.
Microbial Decomposer Clock. Source. Allwood et al (22).
Examining the microbial ecosystem using soil or dust properties around the crime scene, bacterial and fungal spores, as well as plant substances could also be a highly useful forensic tool for predicting the exact geographical location and even the time of a specific incident.
Following standard collection and preservation of this evidence, different microbial DNA analyses would clarify the whole prokaryotic and eukaryotic community (23-25).
Determining the Time of Death
Postmortem proliferation of the internal microbiome can be used to estimate the time of death, as well as to identify the possible victims and suspects involved in a crime. It should be noted that various factors from temperature and humidity degree to age and gender have different effects on corps even at the very same time and place (26-28).
While Forensic pathologists use core body temperature to estimate post-mortem interval, microbial forensics has been proved to be a useful tool for determining the time of death.
In 2009, a group of forensic scientists faced a crime scene in a closed apartment, where a man’s body was stabbed; they found fungal colonies on carpets and couches. Therefore, they recreated the crime scene by adding those fungi to a mixture of a clean part of the carpet and bovine blood and then compared the results to the original fungal growth back there, which led them to realize that the death had occurred 5 days before finding the corpus (29).
Postmortem Bacterial Translocation
There are several immunological and physical barriers in the human body to prevent bacteria from entering the other parts of the body.
After death, as the intestinal barrier disappears, bacteria enter the bloodstream (to name a few, Bacteroidetes, Enterobacteriaceae, and Staphylococcaceae) and different organs, including the liver, pericardial fluid, and spleen, which are important for bacteriological mortality after death. Bacteria can be detected by cultural methods or real-time quantitative polymerase chain reaction (RT-qPCR).
Blood is the first place where bacteria can be found soon after death. The pericardial fluid remains the most sterile, and the liver remains sterile for up to 5 days after death.
An autopsy can detect bacterial infections after death, including respiratory, administrative, and abdominal infections.
Additionally, post-mortem bacteriology could provide useful data on infection-related mortality when the patient’s history is unavailable (30,31).
Determining the Cause of Death
There are three main problems for isolating and detecting infectious agents, including post-mortem movement, losing vulnerable species, and over-growth of faster-growing organisms. Different microbiological tests can be employed to determine whether a victim is drowned.
Diatoms are useful organisms for classifying unknown drowning victims since they might remain in the liver, bone marrow, and kidneys and could be detected by microscopic examinations and PCR assays.
Examining fecal bacteria such as E. coli and Enterococcus faecalis, which are mostly present in freshwater, is another sensitive tool for identifying drowning cases. Bacteria are present in the drowning medium, thus they can enter different tissues of the body and could be simply detected by growth in special culture media (32).
Bacterioplankton is also used as a marker for assessing drowning. Some studies demonstrated that bacterioplankton capable of proliferating in human blood not only reveals the salinity of water but also prevents bacterial contamination in blood (33,34).
Human Identification
Similar to genetic codes and fingerprints, the human microbiome is also unique. Therefore, it can be applied for serving forensics and security service purposes. In addition, the specific microbial signature that can be found on hair, skin, body fluids, respiratory tract, intestine, genitals, and even feces plays an essential role in developing the personal medicine as each person would receive their personal medications(Figure 3).
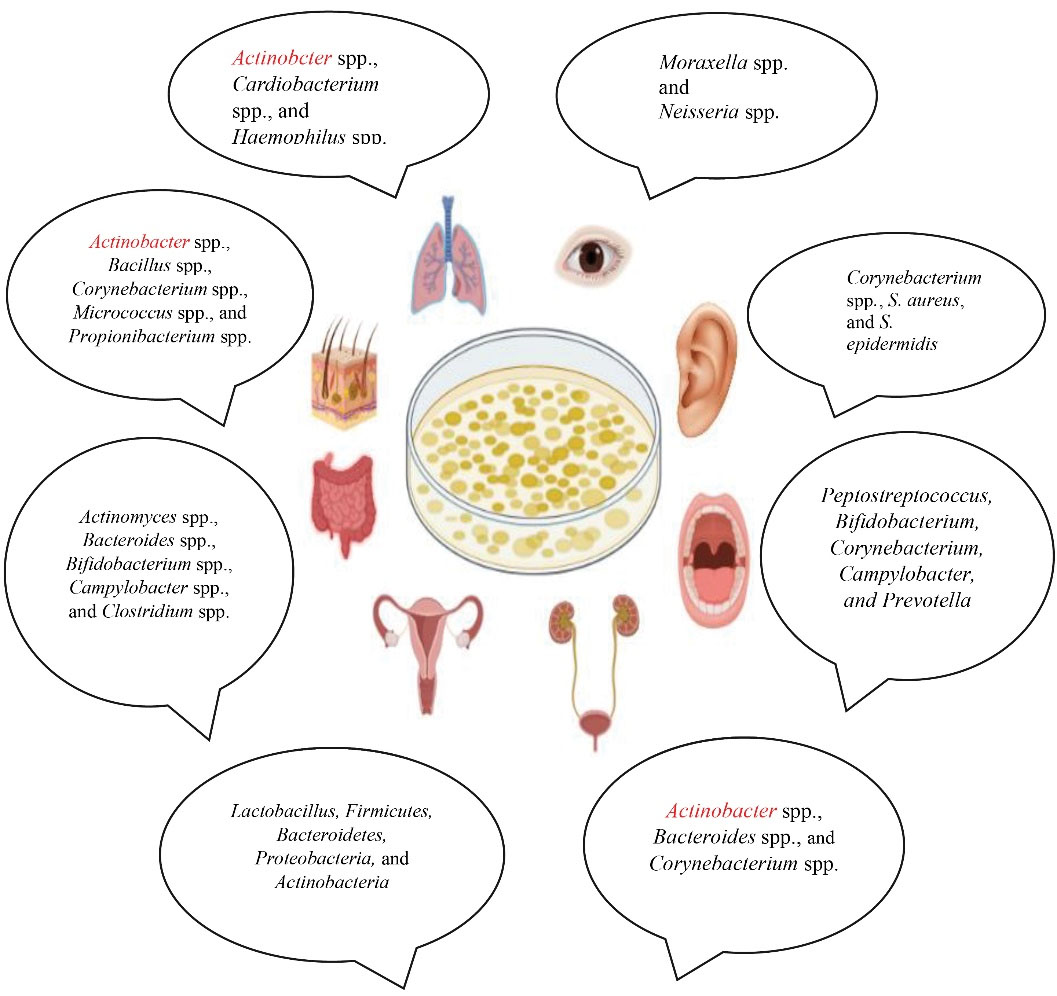
Figure 3.
Human Microbiome Can be Used as Unique Signature to as a Detectable Evidence in Forensic Investigation.
.
Human Microbiome Can be Used as Unique Signature to as a Detectable Evidence in Forensic Investigation.
Human Skin
Human skin is covered with various types of microorganisms, which are remarkably unique to each person, and as a result, this characteristic could be used to identify victims or criminals by tracing the microbial DNA (35). Moreover, it should be noted that in the case of sexual assault, a group of professionals must gather together to inspect all aspects of the case (Figure 4).
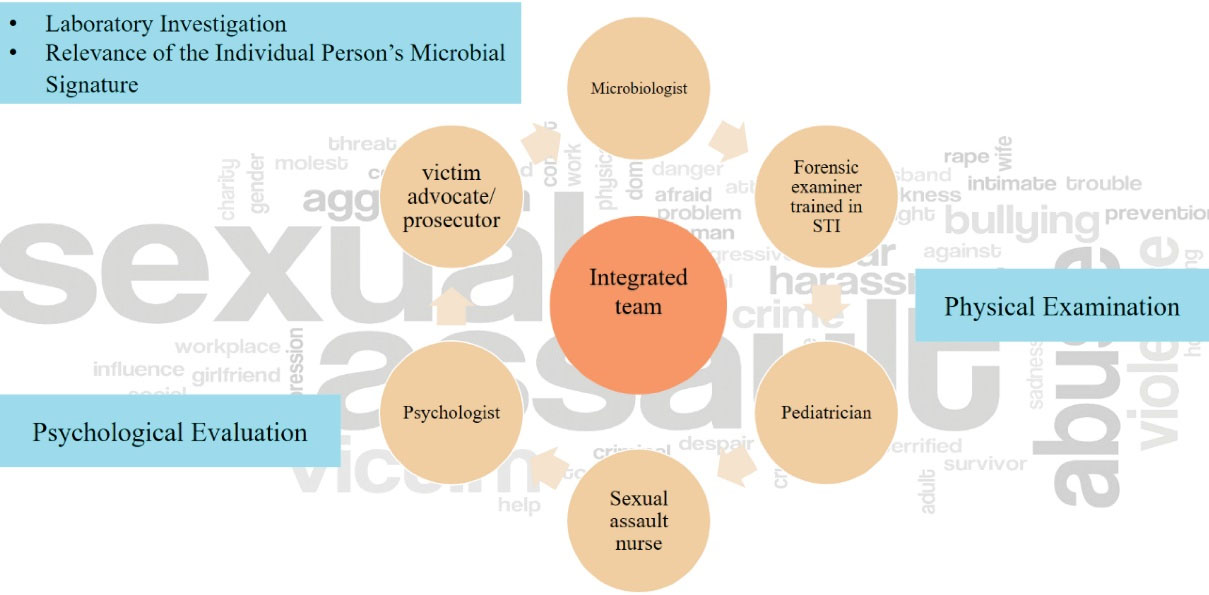
Figure 4.
In a Case of Sexual Assault, a Professional Team Gather Together to Examine and Help the Victim and Find the Possible Offender.
.
In a Case of Sexual Assault, a Professional Team Gather Together to Examine and Help the Victim and Find the Possible Offender.
It is noticeable that when an object hit the body, not only does it burses the skin but also its effects on the microbial community will remain for a while.
This specific sort of evidence is particularly valuable in the case of sexual assaults or any other physical violence, including domestic violence such as child abuse in all sorts of ways (36,37).
Human Hair
Hair is another piece of biological evidence in forensic investigation. Microbial communities can be detected by means of human DNA profiling tests or even special cultural methods.
In line with the mentioned issue, high-resolution optical microscopy, electron microscopy, and scanning electron microscopy or nuclear DNA and mitochondrial DNA tests can be used to investigate the nature of damaged hair fibers. Variations in the severity of hair keratin loss can be considered for identification (38,39).
In addition to microscopic and molecular techniques, other characteristics such as even slight differences between hair colors showing decreasing the pigment could be the sign of probable sunlight exposure (40).
Human Body Fluid
Considering that each person has a specific microbial composition of body fluids, especially semen and saliva, these fluids could be used as detectable evidence at a crime scene which later can be examined by means of the next-generation sequencing technique.
It is a known fact that the vitreous fluid is more resistant to microbial decomposition compared to blood, saliva, or even semen, which makes it suitable to be examined for postmortem ethanol levels (37,41,42).
Based on previous evidence, alcohol consumption is the main contributing reason to traffic accidents, drownings, suicides, and the like. When blood samples are unavailable, vitreous fluid can be occasionally examined as an alternative (43).
Tools
In line with forensic investigation, all sorts of evidence play a role in collecting information, including the people, place, laboratory equipment, forensic tests, and even the time of the crime.
Since genome analysis could help diverge a bioterrorist action and a natural outbreak, DNA typing methods are the main tools for the attribution of pathogens.
Although DNA extraction methods need to improve to obtain more accurate results, After DNA extraction, molecular assays such as sequencing, microarray, pathogenicity array, single-nucleotide polymorphism, 16S rRNA sequencing, antibiotic resistance, and other methods will be performed to achieve the suitable pathogen resolution.
In addition to mentioned DNA-based methods, there are other microbial assays to participate in solving a crime, some of which are presented as follows:
-
Checking the morphology of microorganisms and trying physiological approaches such as phage typing, serotyping, and the like;
-
Conducting isotope analysis to figure out the age and the origin;
-
Examining the left growth medium;
-
Detecting specific local bacteria;
-
Using immunoassays to observe immunological responses.
In the case of using microbial toxins, immunoassays and mass spectrometry alongside DNA- based approaches would help trace evidence and find the origin of the toxin (1,44).
Molecular Typing
During the time that the whole world is dealing with an outbreak, it is vital to detect the infectious agent and its therapy. Therefore, phenotypic and genotypic methods can be employed for identifying and differentiating strains.
Phenotyping Tests
Reproducibility, ease of performance and interpretation, and ability to type all strains are three important characteristics that should be noticed for choosing phenotyping tests. Biotyping methods and antimicrobial susceptibility patterns are easy to perform and can type all strains. Multilocus enzyme electrophoresis is a phenotypic method, which not only has two earlier options but also is an excellent reproducible test. On the other hand, serotyping and phage typing are somehow difficult methods to perform and are not common among all strains (45).
Genotyping Tests
Another kind of microbial test helping the forensic investigations is genotyping methods, which can be listed in three parts as follows:
-
Restriction endonuclease-based methods, including RFLP with and without hybridization;
-
Amplification-based methods such as RAPD, AFLP, and VNTR;
-
Sequence-based methods, including multilocus sequencing typing (MLST), electrophoresis karyotyping, single nucleotide polymorphism (SNP) analysis, and last but not least whole-genome sequencing (46-48).
Matrix-assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
MALDI-TOF MS is a high-performance technology focused on a comparison of the protein fingerprint produced by microbial cells with the reference spectrum database using various algorithms on the species level, which is applied in various clinical identification occasions for both bacteria and fungi. Using this method in forensic investigation has also some challenges, including a vast reliable database and the ability to differentiate related biothreat agents (49,50).
Proteomics approaches can occasionally provide important data that cannot be achieved from DNA-based methods such as MALDI-TOF MS and LC-MS. Although metagenomics has recently become the standard method for identification and classification, it can be used in the criminal justice system (51).
Massively Parallel Sequencing Approach
Metagenomic analysis has mainly concentrated on the 16S rRNA gene, which is inadequate for distinguishing subspecies. In addition, using a single gene target leads to other restrictions, including copy number variation, sequence variability among a single bacterium, and horizontal gene transfer. Therefore, scientists need to increase sampling and extend databases while adding new genomic sequences to advance specificity. To this end, highly developed technology and costly validation are necessary (52-55).
Using multiple phylogenetic markers, especially housekeeping genes (containing conserved and variable regions) has proved to be a suitable way to obtain more information for species resolution (56).
MLST is one of the methods, with the aid of bioinformatics tools and the use of 20 genes, for identifying bacteria (57).
Considering the limitations of these methods, there are other approaches to conduct, including multilocus variable-number tandem-repeat analysis, SNP analysis, and ribosomal MLST (58).
Bioinformatics
Given the growing use of microbial forensic tests in the criminal justice system, optimizing computational approaches for downstream applications is an important challenge, resulting in overcoming bioinformatics. For instance, studies indicated that among different bioinformatics pipelines, including MG-RAST, Mothur, and QIIME2, the last one would be the suitable method for large postmortem microbial communities (59).
The findings of a study demonstrated that using hidSkinPlex as a new enrichment method could help forensic scientists profile the human skin microbiome and as a result, human identification (60).
Validation
Validation is a necessary step in microbial forensics since not only the procedure but also the results can highly affect public health, political decision, and court vote. This process is summarized as follows:
-
Assessing the ability of procedures to obtain reliable results under defined conditions;
-
Rigorously defining the conditions that are required to obtain the results;
-
Determining the limitations of the procedures;
-
Identifying the aspects of the analysis that must be monitored and controlled;
-
Forming the basis for the development of interpretation guidelines to convey the significance of the findings (61).
As stated earlier, microbial forensic findings could be the actual evidence before the law; therefore, the whole procedure should follow a valid paradigm from the moment the forensic team enters the crime scene or face a bio-attack until the moment that the results are submitted to the court.
Moreover, when it comes to bioterrorist attacks, so many governments might take responsibility. It takes at least four sovereign nations to join the international investigation and each will be subject to one critical task as mentioned below (61).
-
Taking military actions;
-
Taking internal police actions;
-
Taking internal regulatory actions;
-
Supporting political actions.
Conclusions
As mentioned before, DNA extraction is an essential part of microbial forensics examinations. Thus, novel proper extraction and sequencing methods to develop reproducibility, sensitivity, and accuracy are crucial targets. In addition, finding a relation between post-mortem gene expression and microbial colonization would be a new area of interest.
Ultimately, almost every identification approaches rely on established databases, which needs to be expanded not only for human microbiome signatures but also for illustration of local microbial patterns in different areas, pollution source tracking, geolocation, Post-mortem interval (PMI), and the like.
Some countries only use microbial forensic examination in special situations like sexual transmitted disease (STD) victims. Considering the widespread applications of this scientific field in solving crimes, especially bio-crimes, different legal systems could use its advantages to establish a proper chain of custody and obtain more accurate results.
Conflict of Interests
The authors declared no conflict of interests related to this paper.
Ethical Approval
Not applicable.
References
- Budowle B, Schutzer SE, Einseln A, Kelley LC, Walsh AC, Smith JA. Public healthBuilding microbial forensics as a response to bioterrorism. Science 2003; 301(5641):1852-3. doi: 10.1126/science.1090083 [Crossref] [ Google Scholar]
- Lee SB, Mills DK, Morse SA, Schutzer SE, Budowle B, Keim P. Education and training in microbial forensics. In: Budowle B, Schutzer S, Morse S, eds. Microbial Forensics. 3rd ed. Academic Press; 2020. p. 473-95. 10.1016/b978-0-12-815379-6.00032-5.
- Ciesielski C, Marianos D, Ou CY, Dumbaugh R, Witte J, Berkelman R. Transmission of human immunodeficiency virus in a dental practice. Ann Intern Med 1992; 116(10):798-805. doi: 10.7326/0003-4819-116-10-798 [Crossref] [ Google Scholar]
- Christopher GW, Cieslak TJ, Pavlin JA, Eitzen EM Jr. Biological warfareA historical perspective. JAMA 1997; 278(5):412-7. [ Google Scholar]
- Grad YH, Lipsitch M, Feldgarden M, Arachchi HM, Cerqueira GC, Fitzgerald M. Genomic epidemiology of the Escherichia coli O104:H4 outbreaks in Europe, 2011. Proc Natl Acad Sci U S A 2012; 109(8):3065-70. doi: 10.1073/pnas.1121491109 [Crossref] [ Google Scholar]
- Keim PS, Budowle B, Ravel J. Microbial forensic investigation of the anthrax-letter attacks. In: Budowle B, Schutzer SE, Breeze RG, Keim PS, Morse SA, eds. Microbial Forensics. 2nd ed. San Diego: Academic Press; 2011. p. 15-25. 10.1016/b978-0-12-382006-8.00002-5.
- National Research Council (NRC). Review of the Scientific Approaches Used During the FBI’s Investigation of the 2001 Anthrax Letters. NRC; 2011.
- Price EP, Seymour ML, Sarovich DS, Latham J, Wolken SR, Mason J. Molecular epidemiologic investigation of an anthrax outbreak among heroin users, Europe. Emerg Infect Dis 2012; 18(8):1307-13. doi: 10.3201/eid1808.111343 [Crossref] [ Google Scholar]
- Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 2014; 345(6202):1369-72. doi: 10.1126/science.1259657 [Crossref] [ Google Scholar]
- Carus WS. Bioterrorism and Biocrimes: The Illicit Use of Biological Agents Since 1900. Washington, DC: National Defense University; 2001.
- Centers for Disease Control and Prevention. Steps of an Outbreak Investigation. Available from: https://www.cdc.gov/csels/dsepd/ss1978/lesson6/section2.html. Accessed September 15, 2016.
- Treadwell TA, Koo D, Kuker K, Khan AS. Epidemiologic clues to bioterrorism. Public Health Rep 2003; 118(2):92-8. doi: 10.1093/phr/118.2.92 [Crossref] [ Google Scholar]
- Horrox R. The Black Death. Manchester University Press; 2013.
- Davis J, Johnson-Winegar A. The Anthrax Terror. DOD’s Number-One Biological Threat. Alabama: Maxwell Air Force Base; 2000.
- McEwen SA, Wilson TM, Ashford DA, Heegaard ED, Kournikakis B. Microbial forensics for natural and intentional incidents of infectious disease involving animals. Rev Sci Tech 2006; 25(1):329-39. [ Google Scholar]
- Thompson J, Clark G. Rabbit calicivirus disease now established in New Zealand. Surveillance 1997; 24(4):5-6. [ Google Scholar]
- Török TJ, Tauxe RV, Wise RP, Livengood JR, Sokolow R, Mauvais S. A large community outbreak of salmonellosis caused by intentional contamination of restaurant salad bars. JAMA 1997; 278(5):389-95. doi: 10.1001/jama.1997.03550050051033 [Crossref] [ Google Scholar]
- Centers for Disease Control and Prevention. Emergency Preparedness and Response. Available from: https://emergency.cdc.gov/agent/agentlist-category.asp. Accessed April 4, 2018.
- Hawksworth DL, Wiltshire PE. Forensic mycology: the use of fungi in criminal investigations. Forensic Sci Int 2011; 206(1-3):1-11. doi: 10.1016/j.forsciint.2010.06.012 [Crossref] [ Google Scholar]
- Metcalf JL, Wegener Parfrey L, Gonzalez A, Lauber CL, Knights D, Ackermann G. A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. Elife 2013; 2:e01104. doi: 10.7554/eLife.01104 [Crossref] [ Google Scholar]
- Metcalf Lab. Microbial Clock of Death. Available from: http://www.jessicalmetcalf.com/outreach-4#outreach.
- Allwood JS, Fierer N, Dunn RR. The future of environmental DNA in forensic science. Appl Environ Microbiol 2020; 86(2):e01504-19. doi: 10.1128/aem.01504-19 [Crossref] [ Google Scholar]
- Allwood JS, Fierer N, Dunn RR, Breen M, Reich BJ, Laber EB. Use of standardized bioinformatics for the analysis of fungal DNA signatures applied to sample provenance. Forensic Sci Int 2020; 310:110250. doi: 10.1016/j.forsciint.2020.110250 [Crossref] [ Google Scholar]
- Velsko SP. Inferential validation and evidence interpretation. In: Budowle B, Schutzer S, Morse S, eds. Microbial Forensics. 3rd ed. Academic Press; 2020. p. 361-80. 10.1016/b978-0-12-815379-6.00024-6.
- Javan GT, Finley SJ, Can I, Wilkinson JE, Hanson JD, Tarone AM. Human thanatomicrobiome succession and time since death. Sci Rep 2016; 6:29598. doi: 10.1038/srep29598 [Crossref] [ Google Scholar]
- Deel H, Bucheli S, Belk A, Ogden S, Lynne A, Carter DO, et al. Using microbiome tools for estimating the postmortem interval. In: Budowle B, Schutzer S, Morse S, eds. Microbial Forensics. 3rd ed. Academic Press; 2020. p. 171-91. 10.1016/b978-0-12-815379-6.00012-x.
- Hauther KA, Cobaugh KL, Jantz LM, Sparer TE, DeBruyn JM. Estimating time since death from postmortem human gut microbial communities. J Forensic Sci 2015; 60(5):1234-40. doi: 10.1111/1556-4029.12828 [Crossref] [ Google Scholar]
- Hawksworth DL. Final Report on Mycological Findings Associated with Operation Lynx. Dundee: Report for Tayside Police; 2009.
- Palmiere C, Egger C, Prod’Hom G, Greub G. Bacterial translocation and sample contamination in postmortem microbiological analyses. J Forensic Sci 2016; 61(2):367-74. doi: 10.1111/1556-4029.12991 [Crossref] [ Google Scholar]
- Tuomisto S, Karhunen PJ, Vuento R, Aittoniemi J, Pessi T. Evaluation of postmortem bacterial migration using culturing and real-time quantitative PCR. J Forensic Sci 2013; 58(4):910-6. doi: 10.1111/1556-4029.12124 [Crossref] [ Google Scholar]
- Lucci A, Campobasso CP, Cirnelli A, Lorenzini G. A promising microbiological test for the diagnosis of drowning. Forensic Sci Int 2008; 182(1-3):20-6. doi: 10.1016/j.forsciint.2008.09.004 [Crossref] [ Google Scholar]
- Kakizaki E, Kozawa S, Imamura N, Uchiyama T, Nishida S, Sakai M. Detection of marine and freshwater bacterioplankton in immersed victims: post-mortem bacterial invasion does not readily occur. Forensic Sci Int 2011; 211(1-3):9-18. doi: 10.1016/j.forsciint.2011.03.036 [Crossref] [ Google Scholar]
- Kakizaki E, Kozawa S, Matsuda H, Muraoka E, Uchiyama T, Sakai M. In vitro study of possible microbial indicators for drowning: salinity and types of bacterioplankton proliferating in blood. Forensic Sci Int 2011; 204(1-3):80-7. doi: 10.1016/j.forsciint.2010.05.006 [Crossref] [ Google Scholar]
- Schmedes SE, Woerner A, Budowle B. Forensic human identification using skin microbiome genetic signatures. In: Budowle B, Schutzer S, Morse S, eds. Microbial Forensics. 3rd ed. Academic Press; 2020. p. 155-69. 10.1016/b978-0-12-815379-6.00011-8.
- Rahimi M, Heng NC, Kieser JA, Tompkins GR. Genotypic comparison of bacteria recovered from human bite marks and teeth using arbitrarily primed PCR. J Appl Microbiol 2005; 99(5):1265-70. doi: 10.1111/j.1365-2672.2005.02703.x [Crossref] [ Google Scholar]
- Oliveira M, Amorim A. Microbial forensics: new breakthroughs and future prospects. Appl Microbiol Biotechnol 2018; 102(24):10377-91. doi: 10.1007/s00253-018-9414-6 [Crossref] [ Google Scholar]
- Wilson AS, Dodson HI, Janaway RC, Pollard AM, Tobin DJ. Selective biodegradation in hair shafts derived from archaeological, forensic and experimental contexts. Br J Dermatol 2007; 157(3):450-7. doi: 10.1111/j.1365-2133.2007.07973.x [Crossref] [ Google Scholar]
- Linch CA. Degeneration of nuclei and mitochondria in human hairs. J Forensic Sci 2009; 54(2):346-9. doi: 10.1111/j.1556-4029.2008.00972.x [Crossref] [ Google Scholar]
- Deedrick DW, Koch SL. Microscopy of hair part 1: a practical guide and manual for human hairs. Forensic Science Communications 2004;6(1).
- Harper DR. A comparative study of the microbiological contamination of postmortem blood and vitreous humour samples taken for ethanol determination. Forensic Sci Int 1989; 43(1):37-44. doi: 10.1016/0379-0738(89)90120-5 [Crossref] [ Google Scholar]
- Yang Y, Xie B, Yan J. Application of next-generation sequencing technology in forensic science. Genomics Proteomics Bioinformatics 2014; 12(5):190-7. doi: 10.1016/j.gpb.2014.09.001 [Crossref] [ Google Scholar]
- Savini F, Tartaglia A, Coccia L, Palestini D, D’Ovidio C, de Grazia U. Ethanol determination in post-mortem samples: correlation between blood and vitreous humor concentration. Molecules 2020; 25(12):2724. doi: 10.3390/molecules25122724 [Crossref] [ Google Scholar]
- Budowle B, Wilson MR, Burans JP, Breeze RG, Chakraborty R. Microbial forensics. In: Breeze RG, Budowle B, Schutzer SE, eds. Microbial Forensics. Burlington: Academic Press; 2005. p. 1-25. 10.1016/b978-012088483-4/50004-4.
- Tenover FC, Arbeit RD, Goering RV. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologistsMolecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol 1997; 18(6):426-39. doi: 10.1086/647644 [Crossref] [ Google Scholar]
- Metcalf JL, Xu ZZ, Weiss S, Lax S, Van Treuren W, Hyde ER. Microbial community assembly and metabolic function during mammalian corpse decomposition. Science 2016; 351(6269):158-62. doi: 10.1126/science.aad2646 [Crossref] [ Google Scholar]
- Cobaugh KL, Schaeffer SM, DeBruyn JM. Functional and structural succession of soil microbial communities below decomposing human cadavers. PLoS One 2015; 10(6):e0130201. doi: 10.1371/journal.pone.0130201 [Crossref] [ Google Scholar]
- Finley SJ, Pechal JL, Benbow ME, Robertson BK, Javan GT. Microbial signatures of cadaver gravesoil during decomposition. Microb Ecol 2016; 71(3):524-9. doi: 10.1007/s00248-015-0725-1 [Crossref] [ Google Scholar]
- Wieser A, Schneider L, Jung J, Schubert S. MALDI-TOF MS in microbiological diagnostics-identification of microorganisms and beyond (mini review). Appl Microbiol Biotechnol 2012; 93(3):965-74. doi: 10.1007/s00253-011-3783-4 [Crossref] [ Google Scholar]
- Duriez E, Armengaud J, Fenaille F, Ezan E. Mass spectrometry for the detection of bioterrorism agents: from environmental to clinical applications. J Mass Spectrom 2016; 51(3):183-99. doi: 10.1002/jms.3747 [Crossref] [ Google Scholar]
- Merkley ED,. Applications in Forensic Proteomics: Protein Identification and Profiling. American Chemical Society; 2019.
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol 2000; 66(4):1328-33. doi: 10.1128/aem.66.4.1328-1333.2000 [Crossref] [ Google Scholar]
- Wang Y, Zhang Z, Ramanan N. The actinomycete Thermobisporabispora contains two distinct types of transcriptionally active 16S rRNA genes. J Bacteriol 1997; 179(10):3270-6. doi: 10.1128/jb.179.10.3270-3276.1997 [Crossref] [ Google Scholar]
- Asai T, Zaporojets D, Squires C, Squires CL. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci U S A 1999; 96(5):1971-6. doi: 10.1073/pnas.96.5.1971 [Crossref] [ Google Scholar]
- Schouls LM, Schot CS, Jacobs JA. Horizontal transfer of segments of the 16S rRNA genes between species of the Streptococcus anginosus group. J Bacteriol 2003; 185(24):7241-6. doi: 10.1128/jb.185.24.7241-7246.2003 [Crossref] [ Google Scholar]
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 1998; 95(6):3140-5. doi: 10.1073/pnas.95.6.3140 [Crossref] [ Google Scholar]
- Keim P, Van Ert MN, Pearson T, Vogler AJ, Huynh LY, Wagner DM. Anthrax molecular epidemiology and forensics: using the appropriate marker for different evolutionary scales. Infect Genet Evol 2004; 4(3):205-13. doi: 10.1016/j.meegid.2004.02.005 [Crossref] [ Google Scholar]
- Jolley KA, Bliss CM, Bennett JS, Bratcher HB, Brehony C, Colles FM. Ribosomal multilocus sequence typing: universal characterization of bacteria from domain to strain. Microbiology (Reading) 2012; 158(Pt 4):1005-15. doi: 10.1099/mic.0.055459-0 [Crossref] [ Google Scholar]
- Kaszubinski SF, Pechal JL, Schmidt CJ, Jordan HR, Benbow ME, Meek MH. Evaluating bioinformatic pipeline performance for forensic microbiome analysis. J Forensic Sci 2020; 65(2):513-25. doi: 10.1111/1556-4029.14213 [Crossref] [ Google Scholar]
- Schmedes SE, Woerner AE, Novroski NMM, Wendt FR, King JL, Stephens KM. Targeted sequencing of clade-specific markers from skin microbiomes for forensic human identification. Forensic Sci Int Genet 2018; 32:50-61. doi: 10.1016/j.fsigen.2017.10.004 [Crossref] [ Google Scholar]
- Budowle B, Schutzer SE, Morse SA, Martinez KF, Chakraborty R, Marrone BL. Criteria for validation of methods in microbial forensics. Appl Environ Microbiol 2008; 74(18):5599-607. doi: 10.1128/aem.00966-08 [Crossref] [ Google Scholar]
- Bidwell CA, Murch R. Use of microbial forensics data in scientific, legal, and policy contexts. In: Budowle B, Schutzer S, Morse S, eds. Microbial Forensics. 3rd ed. Academic Press; 2020. p. 393-404. 10.1016/b978-0-12-815379-6.00026-x.