Avicenna Journal of Clinical Microbiology and Infection. 8(4):135-138.
doi: 10.34172/ajcmi.2021.25
Original Article
Anti-Helicobacter pylori Activity of New Derivatives of 1, 3,4-Oxadiazole: In Silico Study
Yasin SarveAhrabi * 
Author information:
Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran
*
Corresponding author: Yasin SarveAhrabi, Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran, Email:
yasin.ahrabi2016@gmail.com
Abstract
Background: The growing spread of drug resistance in Helicobacter pylori has caused concern. Urease is one of the most important enzymes associated with H. pylori activity. Oxadiazoles have a wide range of inhibitory activities. The aim of this study was to investigate new oxadiazole compounds as urease inhibitors of H. pylori.
Methods: The synthesized compounds were reused as ligands in the previous study, and the initial structure of the compounds was optimized by the Molecular Mechanics Models method. Then, the compounds were evaluated as inhibitors on the active site of the urease enzyme by AutoDock Vina software, and the output results were analyzed and evaluated using soft Discovery Studio software.
Results: All compounds, especially (4c) with flour groups, exhibited powerful inhibitory activity against the urease enzyme of H. pylori.
Conclusions: The present findings indicated the inhibitory potential of the novel synthetic 1, 3, 4-oxadiazole compounds.
Keywords: Helicobacter pylori, Urease, Oxadiazoles, Molecular docking simulation
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Helicobacter pylori is a gram-negative, spiral-shaped bacterium of the genus Epsilonproteobacteria (1). More than 50% of the world’s population is infected with the bacterium, and the infection usually appears in the early years of life (2). The spiral shape allows this bacterium to enter between the mucous layers that cover the stomach. The bacterium breaks down urea molecules, a chemical that produces ammonia and carbon dioxide. Ammonia forms a sheath around the bacteria that protects it against stomach acid. Therefore, the production of large amounts of urease enzyme is essential for the survival and pathogenicity of H. pylori (3), which has been identified by the World Health Organization as a human carcinogen of group 1. This bacterium causes severe gastritis and peptic ulcers in all infected people (4). The prevalence of H. pylori varies based on race, social class, socioeconomic status, health, and age (5). Today, there are several diagnostic and therapeutic methods for H. pylori, there is also treatment resistance that calls for the introduction of new drug combinations (6). In a study conducted in Iran, resistance to metronidazole was very high (57.4%), which is almost consistent with the results of other Asian countries (46.6%); however, the average resistance in Iran is lower than that in African countries (97.55%). Furthermore, the average resistance to ciprofloxacin in Iran is 18.5% due to the small number of studies (7). The enzyme urease has a molecular weight of 540 kDa and consists of two large subunits: UreA with 26.50 kDa and UreB with 61 kDa with a gene size of 1.70 kPa. Urease is a nickel metallic enzyme that hydrolyzes the conversion of urea to ammonium and carbon dioxide. H. pylori activated urease depends on the presence of UreA/B gene structures to form the 550 kDa holoenzyme while UreIE/F subgenes and UreB/I/G genes are required for high expression of urease activity and bacterial establishment in the stomach, respectively.
UreB is the most effective and common immunogen of all H. pylori strains that can elicit a protective immune response in the body against H. pylori (8). Oxadiazole derivatives have a wide range of biological activities including inhibitory properties (9). The increasing need to produce pharmaceutical compounds and replace compounds with resistant drugs has become a necessity; therefore, the purpose of this study was to investigate the molecular docking studies of oxadiazole compounds as potential urease inhibitors of H. pylori.
Methods
Ligand Preparation
Our previously synthesized compounds were reused (10). The initial structure of the compounds was plotted by ChemDraw Pro 12.0 software, then the crystallographic structure of the compounds was drawn using Chem3D 17.0 software, and finally optimized with the Molecular Mechanics Models in kcal.mol-1 command (11). The total energy of each compound after optimization is specified and listed in Figure 1.
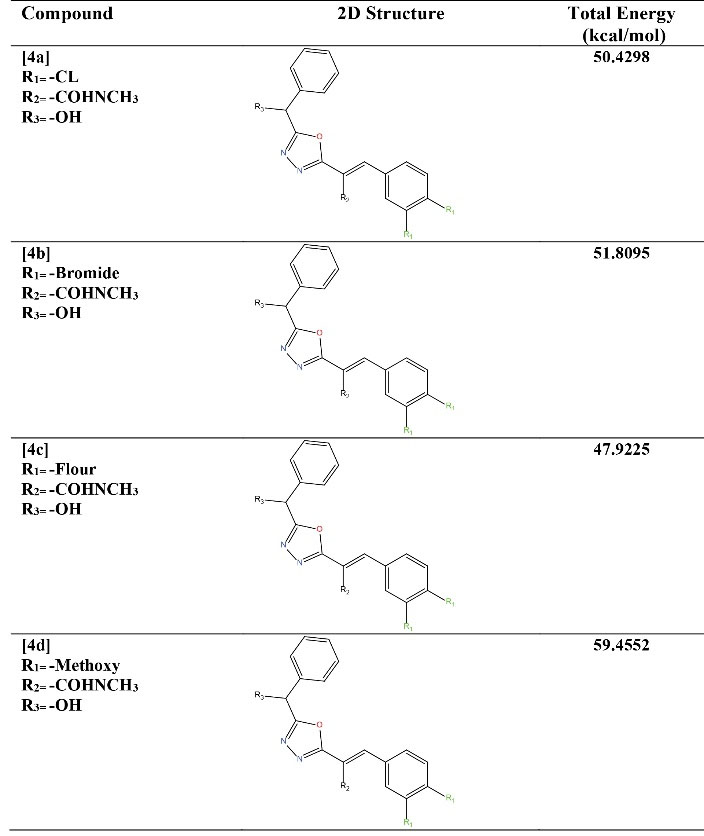
Figure 1.
2D Structural and Total Energy of New Derivatives of 1, 3, 4-Oxadiazoles (10). Note. 2D: Two dimensional.
.
2D Structural and Total Energy of New Derivatives of 1, 3, 4-Oxadiazoles (10). Note. 2D: Two dimensional.
Protein Preparation
Crystallized structure of the urea enzyme of H. pylori was downloaded from the protein database with code (PDB ID: 1e9z) and the resolution of 3 angstroms (Å). To ensure the structure’s health, its crystallography was observed by Visual Molecular Dynamics software. Further, the second structure of the protein was examined through Chimera software.
Molecular Docking
The docking process of compounds 4a-4d to the urease binding site was performed using AutoDock Vina software and Discovery Studio 4.5. Polar hydrogens were added to the compounds and proteins, then the partial charge of the compounds was added, and the partial charge of the protein was added by the Kollman method. Finally, ligand and junction interactions were investigated and analyzed by Discovery Studio 4.5 Client software. All docking calculations were performed using the genetic optimization algorithm and Lamarck traits, which are configured as follows: the maximum number of energy assessments was 25 000 000, and an initial population of 150 was randomly assigned. In addition, the maximum number of generations, the mutation rate, and the crossover rate were 27 000, 0.02, and 0.8, respectively, along with an elitism value. For local search, the so-called Solis algorithm was used with a maximum of 1000 repetitions per search. This process was performed by considering the protein and the ligand as inflexible and flexible, respectively. Grid box with dimensions 120 × 120 × 120 which includes the whole protein, the distance of grade 1 Å, and other parameters (X-Center: 120.00, Y-Center: 100.00, Z-Center: 66.00) was considered, and the best conformity with the lowest amount of binding energy was selected as a result after docking (12).
Results
According to Table 1, all affinities of the compounds were calculated, and the best compounds with low ΔG (-ΔG) were selected to continue experiments and investigate chemical interactions. As demonstrated in Figure 2, the highest amount of hydrogen bonds was related to the 4c compound with the amino acid serine: 567, glutamic acid: 371, threonine: 374, and glutamine: 378, among which the compound bonds were the case. Regarding the 4d compound, the highest amount of carbon-hydrogen bonds was related to Lysine: 445 and Glutamine: 471.
Table 1.
AutoDock Vina Results of 1, 3, 4-Oxadiazole Compounds (4a-4d)
|
No.
|
Affinity
(kcal/mol)
|
Carbon-Hydrogen Bond
|
Pi-Alkyl
Bond
|
Pi-Anion
Bond
|
Hydrogen
Bond
|
Halogen Bond
|
Pi-Cation
Bond
|
| 4a |
-7.3 |
Asparagine: 111 |
Alanine: 94
Lysine: 92 |
Glutamic acid: 64 |
- |
- |
- |
| 4b |
-6.8 |
- |
- |
- |
- |
- |
- |
| 4c |
-8.3 |
- |
Alanine: 563
Valine: 560 |
- |
Serine: 567
Glutamic acid: 371
Threonine: 374
Glutamine: 378 |
Valine: 444 |
Arginine: 375 |
| 4d |
-7.0 |
Lysine: 445
Glutamine: 471 |
Alanine: 16
Valine: 33
Valine: 473 |
Isoleucine: 568 |
- |
- |
- |
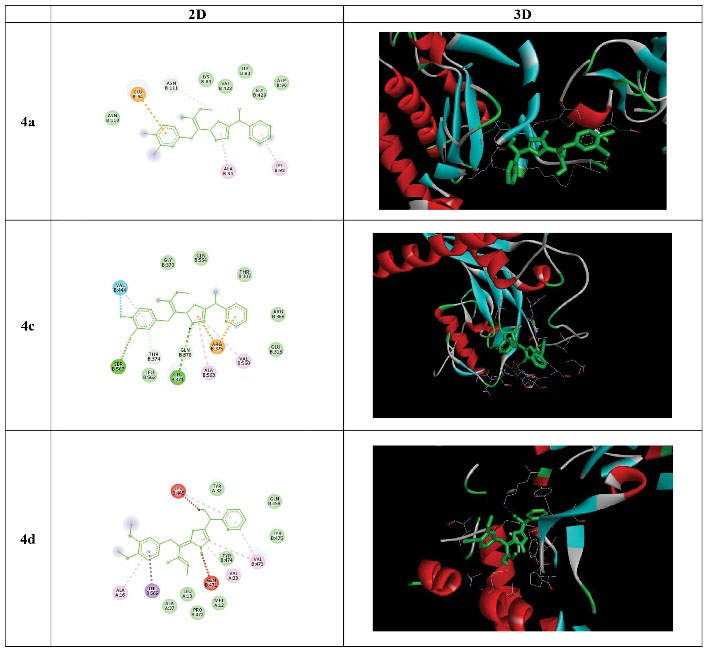
Figure 2.
2D and 3D Results of AutoDock Vina. Note. 2D: Two dimensional; 3D: Three dimensional.
.
2D and 3D Results of AutoDock Vina. Note. 2D: Two dimensional; 3D: Three dimensional.
Discussion
Compounds with 1,3,4-oxadiazole cores have occupied a specific place in medicinal and synthetic chemistry because of their extensive range of biological activities such as antibacterial, antifungal, anti-inflammatory, antiviral, anticancer, enzyme inhibitor, anticonvulsant, and anti-diabetic properties (13). These properties make it a desirable medicinal backbone that can be used to construct biologically-useful molecules. This study evaluated the in silico effects of 4 syntheses of 1, 3, 4-oxadiazole derivatives against the human pathogen H. pylori. Drug resistance of H. pylori has been increasing over the past few decades (14). According to a study by Moran-Gilad et al, H. pylori resistance rate was 54%, 31%, 10%, 4%, and 2% to clarithromycin, metronidazole, amoxicillin, rifampicin, and levofloxacin, respectively (15). Resistance was found to be an alarm to increase drug resistance to treatment. Therefore, compounds with central nuclei of 1, 3, 4-oxadiazole can be a good alternative to existing drugs. In the present study, compound 4c with the flour group appeared with a promising inhibitory activity that can be used as an inhibitor of this compound in the synthesis of anti-H. pylori drugs. In a study conducted by Khan et al, oxadiazole derivatives were evaluated as urease inhibitors, and the results showed that compound 6a had the lowest Ki among the series, while compounds 6d, 6e, 6g, and 6i were subsequently found to have significant Ki values after 6a. Molecular docking has supported their inhibition types and structure activity-relationship (16). In another study, Mao et al synthesized14 metronidazole derivatives (compounds 3a–f and 4b–h) by coupling the metronidazole and salicylic acid derivatives. The docking results revealed that the AutoDock 4.0 program could explain the inhibitory activities of compound 4g (bind Cl group in structure) against H. pylori urease (17). The results also indicated that group Cl could inhibit the H. pylori urease enzyme, but results of the present study indicated that the flour group can inhibit the urease enzyme. Moreover, in their study, Kataria and Khatkar synthesized morin analogous with oxadiazole group as H. pylori urease inhibitors. Among their synthesized compounds, N-(2-chlorophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene) thiourea (M2b) and N-(4-bromophenyl)-N-((4E)-2-(2,4-dihydroxyphenyl)-3,5,7-trihydroxy-4H-chromen-4-ylidene) thiourea (M2i) were found to be most potent urease inhibitors in silico. The results of this study confirmed the effects of chlorine-containing compounds on the inhibition of urease activity of H. pylori (18). The results of the above-mentioned studies are consistent with the results of the present study so that in many studies the main role of the 1, 3, 4-oxadiazole ring has been determined. Further, the combination of 4a with chloride group and 4d with methoxy group showed inhibitory activity so that they had fewer bonding connections than the 4C combination. Moreover, the simple workup, tall abdicates, and brief response times make the strategy a valuable expansion for planning advanced pharmaceutical synthetics.
Conclusions
Helicobacter pylori urease enzyme is one of the most important enzymes associated with bacterial activity, so the inhibition of this enzyme will kill bacteria. Alternative drug structures are a basic and important need in the treatment of pathogenic strains of bacteria. Hence, the need to make and synthesize alternative drugs has increased gradually. Newly synthesized structures containing the central nucleus of 1, 3, 4-oxadiazole can be used for a variety of biological activities. It appears that oxadiazole subordinates will be a supportive structure for conceivable improvement of unused drugs, but this result must be confirmed by other broad clinical trials that will be a portion of our future plans.
Acknowledgments
The author appreciates all professors who participated voluntarily in this investigation.
Conflict of Interests
None.
References
- Gilbreath JJ, Cody WL, Merrell DS, Hendrixson DR. Change is good: variations in common biological mechanisms in the epsilonproteobacterial genera Campylobacter and Helicobacter. Microbiol Mol Biol Rev 2011; 75(1):84-132. doi: 10.1128/mmbr.00035-10 [Crossref] [ Google Scholar]
- Hoi HT. H pylori bacteria–cause of peptic ulcer and some treatments. J Crit Rev 2020; 7(14):887-90. [ Google Scholar]
- Keogan D. The Development of Metallohydroxamates as Novel Anti-Bacterial and Anti-Leishmanial Agents [thesis]. Royal College of Surgeons in Ireland; 2018.
- Butt J, Epplein M. Helicobacter pylori and colorectal cancer-a bacterium going abroad?. PLoS Pathog 2019; 15(8):e1007861. doi: 10.1371/journal.ppat.1007861 [Crossref] [ Google Scholar]
- Kakooza S. Prevalence and factors associated with Helicobacter pylori among adults between 18 and 40 years at Butembe Health Centre III Kyankwanzi district, Uganda. Stud J Health Res Afr 2021; 2(6):7. doi: 10.51168/sjhrafrica.v2i6.37 [Crossref] [ Google Scholar]
- Saleem N, Howden CW. Update on the management of Helicobacter pylori infection. Curr Treat Options Gastroenterol 2020:1-12. doi: 10.1007/s11938-020-00300-3 [Crossref]
- Bakhshi S, Ghazvini K, Beheshti A, Ahadi M, Sheykhi M. Review of antibiotic resistance of Helicobacter pylori in Iran and the world. Med J Mashhad Univ Med Sci 2017; 60(4):648-61. doi: 10.22038/mjms.2017.10191.[Persian] [Crossref] [ Google Scholar]
- Nasr-Esfahani M, Doosti A, Sazegar H. Evaluation of the immune response against Helicobacter pylori; in Infused BALB/c Mice by pcDNA31(+)-ureA. Folia Med (Plovdiv) 2020; 62(1):37-45. doi: 10.3897/folmed.62.e47932 [Crossref] [ Google Scholar]
- Ghameshlouei S, Zarrabi Ahrabi N, Souldozi A, SarveAhrabi Y. Evaluation of the antibacterial and investigation of the molecular docking of new derivatives of 1,3,4-oxadiazole as inhibitors of quorum sensing system in the human pathogen Pseudomonas aeruginosa. Avicenna J Clin Microbiol Infect 2021; 8(1):27-33. doi: 10.34172/ajcmi.2021.06 [Crossref] [ Google Scholar]
- Zarrabi Ahrabi N, Souldozi A, SarveAhrabi Y. Synthesis of new three-component derivatives of 1,3,4-oxadiazole and evaluation of their in vitro antibacterial and antifungal properties. Med Lab J 2021; 15(5):13-8. doi: 10.29252/mlj.15.5.13.[Persian] [Crossref] [ Google Scholar]
- Ghameshlouei S, Zarrabi Ahrabi N, SarveAhrabi Y. In vitro and in silico evaluation of biological activity of a new series of oxadiazole compounds against esp gene expression in Enterococcus faecalis biofilm. Gene Cell Tissue 2021; 8(2):e112403. doi: 10.5812/gct.112403 [Crossref] [ Google Scholar]
- Morris GM, Lim-Wilby M. Molecular docking. In: Kukol A, ed. Molecular Modeling of Proteins. Totowa, NJ: Humana Press; 2008. p. 365-82. 10.1007/978-1-59745-177-2_19
- SarveAhrabi Y, Zarrabi Ahrabi N, Souldozi A. Synthesis and antimicrobial evaluation of new series of 1,3,4-oxadiazole containing cinnamic acid derivatives. Avicenna J Clin Microbiol Infect 2021; 8(1):11-6. doi: 10.34172/ajcmi.2021.03 [Crossref] [ Google Scholar]
- Flores-Treviño S, Mendoza-Olazarán S, Bocanegra-Ibarias P, Maldonado-Garza HJ, Garza-González E. Helicobacter pylori drug resistance: therapy changes and challenges. Expert Rev Gastroenterol Hepatol 2018; 12(8):819-27. doi: 10.1080/17474124.2018.1496017 [Crossref] [ Google Scholar]
- Domanovich-Asor T, Motro Y, Khalfin B, Craddock HA, Peretz A, Moran-Gilad J. Genomic analysis of antimicrobial resistance genotype-to-phenotype agreement in Helicobacter pylori. Microorganisms 2020; 9(1):2. doi: 10.3390/microorganisms9010002 [Crossref] [ Google Scholar]
- Khan FA, Rehman AU, Abbasi MA, Afridi SG, Khan A, Lodhi MA. Structural basis of binding and justification for the urease inhibitory activity of acetamide hybrids of N-substituted 1,3,4-oxadiazoles and piperidines. J Mol Struct 2021; 1223:129141. doi: 10.1016/j.molstruc.2020.129141 [Crossref] [ Google Scholar]
- Mao WJ, Lv PC, Shi L, Li HQ, Zhu HL. Synthesis, molecular docking and biological evaluation of metronidazole derivatives as potent Helicobacter pylori urease inhibitors. Bioorg Med Chem 2009; 17(21):7531-6. doi: 10.1016/j.bmc.2009.09.018 [Crossref] [ Google Scholar]
- Kataria R, Khatkar A. Molecular docking, synthesis, kinetics study, structure-activity relationship and ADMET analysis of morin analogous as Helicobacter pylori urease inhibitors. BMC Chem 2019; 13(1):45. doi: 10.1186/s13065-019-0562-2 [Crossref] [ Google Scholar]