Avicenna Journal of Clinical Microbiology and Infection. 8(3):89-93.
doi: 10.34172/ajcmi.2021.16
Original Article
Application of Multiplex PCR for the Identification of Oxacillinase Genes and Determination of Antibiotic Resistance Pattern in Environmental Isolates of Acinetobacter baumannii in ICU
Anahita Farajzadeh 1, Mohsen Mirzaee 2  , Shahram Nanekarani 3
, Shahram Nanekarani 3  , Reza Yari 1, *
, Reza Yari 1, * 
Author information:
1Department of Biology, Borujerd Branch, Islamic Azad University, Borujerd, Iran
2Department of Laboratory Science, Borujerd Branch, Islamic Azad University, Borujerd, Iran
3Department of Animal Science, Borujerd Branch, Islamic Azad University, Borujerd, Iran
Abstract
Background:
Acinetobacter baumannii is a common cause of nosocomial infections. A prominent feature of these bacteria is resistance to carbapenems. This study aimed to identify OXA genes encoding oxacillinase in Acinetobacter baumannii isolates.
Methods: This cross-sectional descriptive study was performed on 25 environmental A. baumannii isolates collected from ICU over 8 months. Definitive identification of isolates was performed by biochemical tests and polymerase chain reaction (PCR) of 16s rRNA gene. Antibiotic susceptibility testing was performed on Müller-Hinton agar medium by disk diffusion and E-test. Antibiogram and multiplex PCR data of beta-lactamase genes were collected and analyzed at a significance level of P<0.05 using SPSS 22.0.
Results: Except for one isolate, all isolates (96%) were sensitive to polymyxin B and 80% of isolates were sensitive to oxacillin. All isolates were sensitive to meropenem, ampicillin/sulbactam, gentamicin, amikacin, piperacillin, and carbenicillin. The results showed that 25 isolates (100%) had OXA-51 gene, 21 isolates (84%) had OXA-58 gene, one isolate (4%) had OXA-24 gene, and none of the isolates contained OXA-23 gene. Only isolate No.10 had three oxacillinase genes simultaneously and it was resistant to oxacillin, polymyxin B, and cephalothin.
Conclusions: The study showed that environmental isolates of ICU do not have pathogenic genes present in the clinical isolates, and how these genes are transferred to the peripheral isolates is an important point that should be studied. Identification of genes encoding carbapenem resistance may help to understand the mechanisms of resistance transfer in A. baumannii. The lack of the OXA-23 gene plays an important role in the susceptibility of isolates to antibiotics and non-emergence of resistant strains.
Keywords: Acinetobacter baumannii, Carbapenems, Antibiotic resistance, Nosocomial infection
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Acinetobacter baumannii is a gram-negative, oxidase-negative, obligate aerobic, non-fermentative, non-motile, and opportunistic coccobacillus that is abundant in soil and water. Bacteria are spread in the hospital environment on wet surfaces (such as mechanical ventilation) and dry surfaces such as human skin, live in the environment for a long time and are easily transmitted among patients. Due to the remarkable clinical properties of this bacterium and its ability to acquire drug resistance and high reproducibility, it is considered as a microorganism threatening patients hospitalized for treatment with antimicrobial drugs. The genus Acinetobacter is divided into two groups: glucose oxidizing species, the most common of which is A. baumannii, and non glucose-oxidizing species, the most prevalent of which is Acinetobacter lwoffiiand Acinetobacter haemolyticus. The most common cause of infections is A. baumannii, which accounts for most Acinetobacter infections in humans (1,2).
Acinetobacter baumannii causes urinary tract infections, ulcers, meningitis, peritonitis, endocarditis, pneumonia, as well as sepsis in ICU, surgery, infection, burn sections. Those receiving broad-spectrum antibiotics, undergoing surgery, and using artificial respiration, as well as patients who have cystic fibrosis and patients with immunodeficiency are at risk for A. baumannii infections. Acinetobacter infections are difficult to treat because the organism is resistant to most antibiotics, such as carbapenems, due to the presence of OXA family genes, and specific treatment should be selected by in vitro susceptibility testing (1-5). A. baumannii is a pathogen with low sensitivity to antibiotics due to the presence and increased expression of efflux pumps, plasmid, and chromosomal beta-lactamase resistance genes, membrane impermeability, drug binding site change, the presence of some drug-altering enzymes, or different cellular functions of mutants, biofilm production, the presence of simple and complex jumping genetic elements such as IS, transposon and integrons for horizontal transfer of genes and changes in the outer coat of bacteria (1-6). Class D beta-lactamases, also known as OXA-type enzymes or oxacillinases, have lower hydrolyzing activity on carbapenems than metallo-beta-lactamases (MBLs); however, if this group of OXA genes are located near insertion sequence or IS, they can increase their expressions under the influence of their strong promoters. The most important enzyme groups of this family are as follows (6-9):
The OXA-58 enzyme group was first found in France in 2003 and is now reported in many countries. This gene makes the bacterium resistant to penicillins and cephalothin and reduces sensitivity to carbapenems and moxalactam. This gene can be horizontally transferred by motile genetic elements such as plasmids, transposons, integrons, and phages and includes subgroups OXA-58, 96, 97, 164 (6-8).
The OXA-23 enzyme group is predominantly plasmid and was first identified in Scotland in 1993 as ARI-1 (A. baumannii resistant to imipenem-1) and then in Turkey, India, France, Germany, Spain, and the United Kingdom. It is originated from A. radioresistance, a symbiotic bacterium that lives in the skin. Bacteria containing these enzymes are resistant to cephalosporins, cefotaxime, oxacillin, cloxacillin, and other penicillins. This bacterium has 17 subgroups and IS elements that are involved in their activities. OXA-23 gene is the most common carbapenem resistance encoding gene which is translocated horizontally and plays a significant role in bacterial carbapenem resistance (6-8).
The OXA-51 enzyme group is mainly chromosome-encoded and was first identified in 2004 in Argentina. The two subgroups are OXA-51 and OXA-69. Members of this group behave as poor penicillinases and weak carbapenemases but are resistant to inhibition by clavulanic acid and tazobactam. This gene is now the identification marker of A. baumannii (6-8).
The OXA-24 enzyme group is transmitted via the plasmid and has a moderate hydrolytic activity against carbapenems. This enzyme is often active against doripenem (a type of carbapenem) and its configuration is similar to OXA-1. The gene was first identified in the chromosome of a carbapenem-resistant A. baumannii clinical strain isolated in Spain in 1997 and then reported in the Netherlands, the United States, England, and France. In strains with the OXA-24 gene, MIC for carbapenem increased up to 4-fold. OXA-24 is often found in combination with OXA-40 and is now found in other Gram-negative bacilli resistant to carbapenem and penicillins, including Pseudomonas aeruginosa, Klebsiella pneumoniae, Stenotrophomonas maltophilia, and Burkholderia (9-11). OXA-24 includes subgroups such as OXA-231, 143, 253, 72, 160, 40 (6-11).
Multiplex polymerase chain reaction (PCR) is a modified PCR method in which several mutations, genes, alleles, and different infectious agents in different variants can be detected simultaneously using several different pairs of primers in one reaction (2,3,12). In the present study, the multiplex PCR method was used to identify 4 important subgroups of OXA-23, OXA-24, OXA-51, OXA-58 in 25 environmental isolates of A. baumannii in the laboratory plate taken from the ICU (equipment, devices, clothing, etc). The antibiotic resistance profile of the isolates was evaluated at a significance level of P< 0.05 using SPSS 22.0.
Materials and Methods
Sample Collection and Culture
Twenty-five non-repetitive samples of bacteria in the plate collected from the ICU (equipment, tools, clothing, patient pillows) were transferred to the research laboratory of cellular and molecular biology over a period of 8 months in compliance with all ethical principles and international conventions.Blood agar and BHB (Brain Heart Infusion Broth) media were used to grow this bacterium. The bacteria were cultured in Müller-Hinton medium and then subjected to Gram staining, biochemical tests such as oxidase, catalase, oxidative-fermentative test, SIM (Sulfide, Indole, Motility medium), hemolysis, hydrolysis of succinylcholine, nitrate reduction, growth at 44°C, and PCR of 16s rRNA gene (3,5,9). To extract plasmid DNA, Plasmid extraction mini kit (Yekta Tajhiz Azma Company, Cat No. FAPDE050) was used according to the instructions.
Antibiogram
Antibiotic susceptibility testing was performed using Kirby-Bauer method and according to The Clinical & Laboratory Standards Institute (CLSI) 2020 standard guidelines. A 0.5 McFarland standard turbidity of bacteria was prepared and spread on Müller-Hinton agar medium (Merck, Germany) (13).The bacterial suspension was densely cultured by sterile cotton swab on Müller-Hinton agar medium. The antibiotic discs were then placed on the surface of the medium with sterile forceps and the culture medium was incubated for 37-24 hours at 37°C. Antibiotic susceptibility pattern was determined using disk diffusion and E-test for cefotaxime (30 µg), carbenicillin (100 µg), amikacin (30 µg), ampicillin/sulbactam (10/10 µg), cephalothin (30 µg), gentamicin (10 µg), meropenem (10 µg), piperacillin (100 µg), oxacillin (1 µg) and polymyxin B (300 µg) provided by Padten Teb Company, Iran. Escherichia coli ATCC 25922 and A. baumannii PTCC 1388 were used as standard strains in antibiogram test (2,3,5,9). Then, the diameter of the growth inhibition zone was accurately measured and the isolates were reported as resistant, semi-sensitive, and sensitive according to CLSI instructions. Statistical analysis was performed using chi-square and Fisher’s exact test in SPSS 22.0 at a significant level of P< 0.05 (2,14).
Primers, PCR, and Electrophoresis
Total DNA extraction was performed by boiling method. In multiplex PCR method, the frequency of four oxacillinase genes OXA-23, 24, 51, 58 was calculated simultaneously for each isolate using a thermocycler (Bio Rad, USA). Each reaction was performed in a volume of 25 μL containing 1X of PCR buffer, 1 U of Taq enzyme, 2 mM of MgCl2, 200 mM of dNTPs (CinnaGen, Iran), 0.2 μM of each primer, 1 μL of sample DNA and ddH2O for adjusting the reaction volume to 25 μL. Then multiplex PCR was done with the same temperature/time program. The primer sequences of the four oxacillinase genes and the 16s rRNA gene are presented in Table 1. A. baumannii ATCC 1388 was used as the standard strain in PCR reactions. PCR was carried out in a thermocycler as follow: Initial denaturation at 94°C for 4 minutes, 30 cycles of 94°C for 60 seconds, 53°C for 40 seconds, 72°C for 50 seconds, and final extension at 72°C for 5 minutes (12). PCR amplicons were electrophoresed (DeNa Gene Tajhiz, Iran) on 1.5% agarose gel, stained with DNA safe stain (2.5 μL/100 mL, Yekta Tajhiz Azma, Iran) and placed under a UV transilluminator.
Table 1.
The Primers Used in This Study (
2,
12,
15)
|
Gene Name
|
(5→'3') Nucleotide seq.
|
Amplicon Size (bp)
|
|
OXA-51
|
F-5'-TAATGCTTTGATCGGCCTTG-3'
R-5'-TGGATTGCACTTCATCTTGC-3' |
353 |
|
OXA-58
|
F-5'-AAGTATTGGGGCTTGTGCTG-3'
R -5'-CCCCTCTGCGCTCTACATAC-3' |
599 |
|
OXA-23
|
F-5'-GATCGGATTGGAGAACCAGA-3'
R -5'-ATTTCTGACCGCATTTCCAT-3' |
501 |
|
OXA-24
|
F-5'-GGTTAGTTGGCCCCCTTAAA-3'
R -5'-AGTTGAGCGAAAAGGGGATT-3' |
246 |
|
16s rRNA*
|
F-5'-AGAGTTTGATCCTGGCTCAG-3'
R -5'-ACGGCTACCTTGTTACGACTT-3' |
1500 |
* The 16s rRNA gene was not involved in the multiplex reaction but was used in separate PCR to molecularly identify the isolates.
The image obtained from electrophoresis was stored and compared with the 100 bp DNA ladder (Cat No. S-5090, Denazist, Iran).
Results
Antibiogram Results
Antibiotic susceptibility testing was performed by disc diffusion method (Figure 1). The antibiogram patterns of the isolates studied with 10 antibiotics are listed in Table 2. The results showed that except for one isolate, all isolates (96%) were sensitive to polymyxin B. Additionally, 80% of isolates were sensitive to oxacillin (PP = 0.03) and all isolates were sensitive to meropenem, ampicillin/sulbactam, gentamicin, piperacillin, and carbenicillin.

Figure 1.
Antibiotic Susceptibility Testing by Disc Diffusion Method.
.
Antibiotic Susceptibility Testing by Disc Diffusion Method.
Table 2.
Antibiogram Profile of the Studied Isolates
|
Antibiotic Name
|
No. (%) of Resistance Pattern of Isolates
|
P
Value
|
|
Sensitive
|
Semisensitive
|
Resistant
|
| OXA |
(80%)20 |
- |
(20%)5 |
0.03* |
| SAM |
(100%)25 |
(0%) 0 |
(0%)0 |
0.01* |
| GM |
(100%) 25 |
(0%)0 |
(0%)0 |
0.01* |
| AN |
(100%) 25 |
(0%) 0 |
(0%)0 |
0.01* |
| MEN |
(100%) 25 |
(0%) 0 |
(0%) 0 |
0.01* |
| CTX |
(96%)24 |
(4%)1 |
(0%)0 |
0.02* |
| CF |
(96%) 24 |
(0%) 0 |
(4%)1 |
0.02* |
| CB |
(100%) 25 |
(0%)0 |
(0%) 0 |
0.01* |
| PIP |
(100%) 25 |
(0%)0 |
(0%)0 |
0.01* |
| PB |
(96%) 24 |
(0%) 0 |
(4%) 1 |
0.01* |
Abbreviations: CTX, cefotaxime; CB, carbenicillin; AN, amikacin; SAM, ampicillin/sulbactam; CF, cephalothin; GM, gentamicin; MEN, meropenem; PIP, piperacillin; OXA, oxacillin; PB, Polymyxin B.
* P < 0.05 and is significant.
Multiplex PCR Results
Multiple PCR test was used to detect the presence of oxacillinase genes OXA-23, 24, 51, 58 according to the above-mentioned PCR program. After gel electrophoresis, normal PCR test was repeated three times to confirm the absence of genes in isolates without genes. The multiplex PCR products of ten isolates in 1.5% agarose gel electrophoresis are shown in Figure 2.
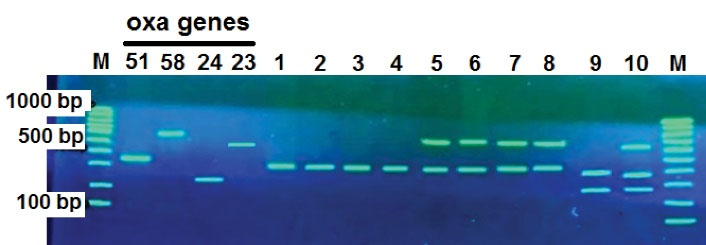
Figure 2.
Multiple PCR Reaction to Investigate the Presence of OXA-58, OXA-23, OXA-24, and OXA-51 Genes on 1.5% Gel Agarose. Left to right M (100 bp marker) and positive control OXA-51 (353 bp), OXA-58 (599 bp), OXA-24 (246 bp) and OXA-23 (501 bp), isolates 1, 2, 3 and 4 have only OXA-51 gene, isolates 5, 6, 7, and 8 have OXA-51 and OXA-58 genes, isolate 9 has OXA-51 and OXA-24 genes. Isolate 10 contained all three genes of OXA-51, OXA-58, and OXA-24, respectively.
.
Multiple PCR Reaction to Investigate the Presence of OXA-58, OXA-23, OXA-24, and OXA-51 Genes on 1.5% Gel Agarose. Left to right M (100 bp marker) and positive control OXA-51 (353 bp), OXA-58 (599 bp), OXA-24 (246 bp) and OXA-23 (501 bp), isolates 1, 2, 3 and 4 have only OXA-51 gene, isolates 5, 6, 7, and 8 have OXA-51 and OXA-58 genes, isolate 9 has OXA-51 and OXA-24 genes. Isolate 10 contained all three genes of OXA-51, OXA-58, and OXA-24, respectively.
Multiplex PCR results showed that 25 isolates (100%) had OXA-51 gene, 21 isolates (84%) had OXA-58 gene, and one isolate (4%) had OXA-24 gene. No isolates contained the OXA-23 gene. Isolates containing two oxacillinase genes were not identified. Isolate No. 10 was the only isolate containing three genes of OXA-24, OXA-51, and OXA-58.
Discussion
Acinetobacter baumannii is increasingly being identified as an important cause of nosocomial infections such as bacteremia and ventilator-associated pneumonia, surgical site infections, toxic sepsis, secondary meningitis, and urinary tract infections especially in ICU, infectious, and burn patients. This bacterium has a high potential for resistance to several classes of antimicrobial agents, which is mainly caused by the production of class D beta-lactamases with carbapenemase activity with the help of jumping genetic elements. The most common resistance-related genes in beta-lactam-resistant isolates include AmpC cephalosporinase genes, OXA-type carbapenemases, MBLs, adeA,S efflux pumps, targeted mutations that alter targets or cellular functions, decreased production of purines such as OprD and integrons which make the use of more antibiotics ineffective and also cause the emergence of strong strains of MDR (multidrug-resistant), XDR (extremely drug-resistant), and PDR (pandrug-resistant) (4,6,9,14-18).
The results of antibiotic susceptibility test showed that except for one isolate (4%), all isolates were sensitive to polymyxin B. Moreover, 80% of the isolates were sensitive to oxacillin (PP = 0.03). All isolates were sensitive to meropenem, ampicillin/sulbactam, gentamicin, piperacillin, and carbenicillin, and fortunately, carbapenem-resistant A. baumannii (CRAB) was not found in this study. These results are different from those of the studies conducted by Tarashi et al (5), Moghadasi et al, Rahmani, Vahhabi et al, and Sarhaddi et al and other studies conducted in different countries due to the clinical nature of the samples in most studies. In the present study, the isolates were derived from non-clinical samples such as equipment and devices used in ICU (2,3,5,15,19-30). In a study conducted by Goudarzi et al, the two groups of clinical and environmental samples were compared and it was revealed that most of the clinical samples were resistant to antibiotics (24). In 2016, Sarhaddi et al found that resistance to antibiotics was more than 95%. In addition, this study demonstrated high prevalence rates of blaVIM, blaIMP, blaTEM, blaADC, OXA-Like23, OXA-Like24, OXA-Like51, OXA-Like58, and ISAba1 genes, which are resistant to beta-lactam antibiotics. These results suggest further studies on the rational administration of drugs in the treatment of infections caused by A. baumannii and the study of the mechanism of resistance transfer from pathogenic isolates to the environment, especially during antibiotic pressure (14,16,18,20,21,24,29).
Four subtypes of oxacillinase enzymes including OXA-23, OXA-24, OXA-51, and OXA-58 have been identified in Acinetobacter. The OXA-51 gene is inherently and chromosomally expressed in A. baumannii. The presence of this gene along with biochemical tests confirms A. baumannii (25,28,30). In our study, the OXA-51 gene was present in all isolates under study, which was consistent with the study of other researchers. The relative frequency of OXA-23 gene in studies conducted in Iran ranges from 4.3 to 100% (2,3,22-27) and in similar studies conducted abroad was reported to range from 31 to 100% (3,15,19,28-30). However, the results of a leading study showed that of the 25 isolates, all were negative for the OXA-23 gene. Scientists attribute the emergence of CRAB strains to the presence of OXA-23 gene (22,27-30), and genetic elements play an important role in its expression (10,11,15,20). According to the results of a study by Zhao et al, the presence of OXA-23 gene is the cause of CRAB phenotype in this bacterium (28) and this may be the reason for the very good sensitivity of our isolates to the antibiotics used because the prevalence of this gene in the present study was 0%. Only one isolate (4%) had the OXA-24 gene, which is consistent with the study conducted by Kooti et al (4.5%) (2). The frequency of this gene in Iran ranges from 0 to 74.1% (3,19,20,22-26) and in other countries, it ranges from 0 to 57.6% (19,21,26,29,30). According to a study conducted by Codjoe and Donkor in 2018, this gene is prevalent in A. baumannii isolates (14). One of the important reasons for this difference is the discrepancy in the type of sample (samples from burn wounds, blood infection, lung, urine, etc. with non-clinical samples used in our study). In the present study, bacterial samples were not obtained from patients but were isolated from the ICU (clothing, equipment, etc.). In our study, 21 isolates (84%) carried the OXA-58 gene, which was consistent with a study by Rahmani et al (3). The frequency of this gene in various studies in Iran has been estimated to range from 0 to 82.1% (2,3,19,20,24,27) and in other countries it ranged from 0 to 60% (19,28-30). The only concern was isolate No. 10, which had three genes, OXA-51, 24 and 58 and was resistant to oxacillin, cephalothin, and polymyxin and relatively resistant to cefotaxime. However, resistance to drugs other than polymyxin is increasing sharply, but resistance to the strongest drug for the treatment of A. baumannii infections needs further epidemiological and molecular studies. Mcr-1 gene plays an important role in resistance to polymyxin B and Colistin (5).
Conclusion
Non-clinical ICU isolates lack important pathogenic genes present in clinical isolates and how these genes are transferred to environmental isolates is an important point that should be studied. Differences in the prevalence of resistance genes and subsequent change in antibiotic resistance patterns in many studies may result from geographical distance, sample size, design of the study, and type of biological sample. Therefore, knowledge of the resistance pattern of each region can help to reduce the incidence of antibiotic resistance, prevent treatment failure, and reduce treatment costs.
Authors’ Contribution
AF: Writing of original draft, investigation. MM: Validation, methodology, and consultation. SN: Data curation and analysis, as well as consultation. RY: Supervision, and editing.
Conflict of Interests
None.
Ethical Approval
Ethical principles were considered in relation to the proposed work and no ethical issues were found to be applied to this research.
Funding/Support
Islamic Azad University of Borujerd Branch provided financial support for this research.
Acknowledgments
The present article was extracted from the MSc thesis (code. 90173) at Islamic Azad University, Borujerd Branch.
References
- Howard A, O’Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 2012; 3(3):243-50. doi: 10.4161/viru.19700 [Crossref] [ Google Scholar]
- Kooti S, Motamedifar M, Sarvari J. Antibiotic resistance profile and distribution of oxacillinase genes among clinical isolates of Acinetobacter baumannii in Shiraz teaching hospitals, 2012 - 2013. Jundishapur J Microbiol 2015; 8(8):e20215. doi: 10.5812/jjm.20215v2 [Crossref] [ Google Scholar]
- Rahmani M, Amirmozafari N, Oshagi M. An investigation of the presence of oxacillinase genes (blaOXA-51, blaOXA-23, blaOXA-58, blaOXA-24) in Acinetobacter baumannii strains isolated from patients in Tehran Imam Khomeini Hospital and Shiraz Namazi Hospital, Iran. Qom Univ Med Sci J 2015; 9(10):55-63. [ Google Scholar]
- Tiwari V, Roy R, Tiwari M. Antimicrobial active herbal compounds against Acinetobacter baumannii and other pathogens. Front Microbiol 2015; 6:618. doi: 10.3389/fmicb.2015.00618 [Crossref] [ Google Scholar]
- Tarashi S, Goudarzi H, Erfanimanesh S, Pormohammad A, Hashemi A. Phenotypic and molecular detection of metallo-beta-lactamase genes among imipenem resistant Pseudomonas aeruginosa and Acinetobacter baumannii strains isolated from patients with burn injuries. Arch Clin Infect Dis 2016; 11(4):e39036. doi: 10.5812/archcid.39036 [Crossref] [ Google Scholar]
- Antunes NT, Fisher JF. Acquired class D β-lactamases. Antibiotics (Basel) 2014; 3(3):398-434. doi: 10.3390/antibiotics3030398 [Crossref] [ Google Scholar]
- Evans BA, Amyes SG. OXA β-lactamases. Clin Microbiol Rev 2014; 27(2):241-63. doi: 10.1128/cmr.00117-13 [Crossref] [ Google Scholar]
- Opazo A, Domínguez M, Bello H, Amyes SG, González-Rocha G. OXA-type carbapenemases in Acinetobacter baumannii in South America. J Infect Dev Ctries 2012; 6(4):311-6. doi: 10.3855/jidc.2310 [Crossref] [ Google Scholar]
- Basatian-Tashkan B, Niakan M, Khaledi M, Afkhami H, Sameni F, Bakhti S. Antibiotic resistance assessment of Acinetobacter baumannii isolates from Tehran hospitals due to the presence of efflux pumps encoding genes (adeA and adeS genes) by molecular method. BMC Res Notes 2020; 13(1):543. doi: 10.1186/s13104-020-05387-6 [Crossref] [ Google Scholar]
- Walther-Rasmussen J, Høiby N. OXA-type carbapenemases. J Antimicrob Chemother 2006; 57(3):373-83. doi: 10.1093/jac/dki482 [Crossref] [ Google Scholar]
- Mlynarcik P, Chalachanova A, Vagnerovă I, Holy O, Zatloukalova S, Kolar M. PCR detection of oxacillinases in bacteria. Microb Drug Resist 2020; 26(9):1023-37. doi: 10.1089/mdr.2019.0330 [Crossref] [ Google Scholar]
- Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006; 27(4):351-3. doi: 10.1016/j.ijantimicag.2006.01.004 [Crossref] [ Google Scholar]
- The Clinical & Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement EP06. Wayne, PA: CLSI; 2020.
- Codjoe FS, Donkor ES. Carbapenem resistance: a review. Med Sci (Basel) 2017; 6(1):1. doi: 10.3390/medsci6010001 [Crossref] [ Google Scholar]
- Joshi PR, Acharya M, Kakshapati T, Leungtongkam U, Thummeepak R, Sitthisak S. Co-existence of bla(OXA-23) and bla(NDM-1) genes of Acinetobacter baumannii isolated from Nepal: antimicrobial resistance and clinical significance. Antimicrob Resist Infect Control 2017; 6:21. doi: 10.1186/s13756-017-0180-5 [Crossref] [ Google Scholar]
- Pagano M, Martins AF, Barth AL. Mobile genetic elements related to carbapenem resistance in Acinetobacter baumannii. Braz J Microbiol 2016; 47(4):785-92. doi: 10.1016/j.bjm.2016.06.005 [Crossref] [ Google Scholar]
- Suwantarat N, Carroll KC. Epidemiology and molecular characterization of multidrug-resistant gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control 2016; 5:15. doi: 10.1186/s13756-016-0115-6 [Crossref] [ Google Scholar]
- Monem S, Furmanek-Blaszk B, Łupkowska A, Kuczyńska-Wiśnik D, Stojowska-Swędrzyńska K, Laskowska E. Mechanisms protecting Acinetobacter baumannii against multiple stresses triggered by the host immune response, antibiotics and outside-host environment. Int J Mol Sci 2020; 21(15):5498. doi: 10.3390/ijms21155498 [Crossref] [ Google Scholar]
- Moghadasi M, Kalantar-Neyestanaki D, Karami-Zarandi M, Rahdar HA, Jasemi S, Feizabadi MM. Investigation of antimicrobial susceptibility patterns and frequency of blaOXA genes in carbapenem resistant Acinetobacter baumannii strains. Sci J Kurdistan Univ Med Sci 2018; 23(4):112-23. [ Google Scholar]
- Sarhaddi N, Dolatabadi S, Amel Jamehdar S. Drug resistance pattern of carbapenem-resistant Acinetobacter baumannii isolated from a referral burn center in northeast of Iran. Med J Mashhad Univ Med Sci 2016; 59(1):1-8. doi: 10.22038/mjms.2016.6981.[Persian] [Crossref] [ Google Scholar]
- Rolain JM, Loucif L, Al-Maslamani M, Elmagboul E, Al-Ansari N, Taj-Aldeen S. Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in Qatar. New Microbes and New Infections 2016; 11:47-51. doi: 10.1016/j.nmni.2016.02.006 [Crossref] [ Google Scholar]
- Pournajaf A, Rajabnia R, Razavi S, Solgi S, Ardebili A, Yaghoubi S. Molecular characterization of carbapenem-resistant Acinetobacter baumannii isolated from pediatric burns patients in an Iranian hospital. Trop J Pharm Res 2018; 17(1):135-41. doi: 10.4314/tjpr.v17i1.19 [Crossref] [ Google Scholar]
- Poirel L, Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect 2006; 12(9):826-36. doi: 10.1111/j.1469-0691.2006.01456.x [Crossref] [ Google Scholar]
- Goudarzi H, Douraghi M, Ghalavand Z, Goudarzi M. Assessment of antibiotic resistance pattern in Acinetobacter bumannii carrying blaOXA type genes isolated from hospitalized patients. Novelty in Biomedicine 2013; 1(2):54-61. doi: 10.22037/nbm.v1i2.5093 [Crossref] [ Google Scholar]
- Mohajeri P, Farahani A, Feizabadi MM, Ketabi H, Abiri R, Najafi F. Antimicrobial susceptibility profiling and genomic diversity of Acinetobacter baumannii isolates: a study in western Iran. Iran J Microbiol 2013; 5(3):195-202. [ Google Scholar]
- Vahhabi A, Hasani A, Ahangarzadeh Rezaee M, Baradaran B, Hasani A, Samadi Kafil H. Carbapenem resistance in Acinetobacter baumannii clinical isolates from northwest Iran: high prevalence of OXA genes in sync. Iran J Microbiol 2021; 13(3):282-93. doi: 10.18502/ijm.v13i3.6388 [Crossref] [ Google Scholar]
- Bahador A, Raoofian R, Farshadzadeh Z, Beitollahi L, Khaledi A, Rahimi S. The prevalence of ISAba 1 and ISAba 4 in Acinetobacter baumannii species of different international clone lineages among patients with burning in Tehran, Iran. Jundishapur J Microbiol 2015; 8(7):e17167. doi: 10.5812/jjm.17167v2 [Crossref] [ Google Scholar]
- Zhao Y, Hu K, Zhang J, Guo Y, Fan X, Wang Y. Outbreak of carbapenem-resistant Acinetobacter baumannii carrying the carbapenemase OXA-23 in ICU of the eastern Heilongjiang province, China. BMC Infect Dis 2019; 19(1):452. doi: 10.1186/s12879-019-4073-5 [Crossref] [ Google Scholar]
- Chang Y, Luan G, Xu Y, Wang Y, Shen M, Zhang C. Characterization of carbapenem-resistant Acinetobacter baumannii isolates in a Chinese teaching hospital. Front Microbiol 2015; 6:910. doi: 10.3389/fmicb.2015.00910 [Crossref] [ Google Scholar]
- Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani SM. Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii isolates in the Gulf Cooperation Council States: dominance of OXA-23-type producers. J Clin Microbiol 2015; 53(3):896-903. doi: 10.1128/jcm.02784-14 [Crossref] [ Google Scholar]