Avicenna Journal of Clinical Microbiology and Infection. 9(3):103-108.
doi: 10.34172/ajcmi.2022.3337
Original Article
Investigating the Inhibitory Effect of Probiotic Lactobacillus Species Isolated From Dairy Products
Azar Rahi 1, *  , Mohammad Hosein Marhamatizadeh 2
, Mohammad Hosein Marhamatizadeh 2 
Author information:
1Department of Pathobiology, School of Public Health, Tehran University of Medical Science. Tehran, Iran
2Department of Food Hygiene, Veterinary Faculty, Kazerun Branch, Islamic Azad University, Kazerun, Iran
Abstract
Background: Lactobacilli are among the most important known probiotic species, and efforts are underway to isolate them from various sources. The beneficial effect of probiotics on health is the main reason for their wide use in dairy products, including yogurt. This research was conducted to isolate and identify Lactobacillus species present in traditional Iranian yogurt.
Methods: Thirty samples of traditional yogurt were collected from different regions of the Gachsaran district south of Iran. Bacterial isolations were performed according to the standard international protocols. Following successive culturing on selective media, obtaining typical colonies, and conducting microscopic examinations, five isolates were selected and analyzed for anti-microbial activities against three bacteria and two pathogenic fungi. To confirm biochemical results, three bacterial isolates were selected for molecular analysis and identified by 16S rRNA sequencing.
Results: Lactobacillus paracasei JCM1171 with 1384 nucleotides and Lactobacillus plantarum subsp. Plantarum.A1 with 1380 nucleotides were isolated based on investigations. The most anti-microbial activities were related to strain A, and the least effects belonged to strain X3. Moreover, among the applied pathogenic microorganisms, Staphylococcus aureus and the fungus, Aspergillus niger had the highest sensitivity.
Conclusion: Probiotic bacteria have highly beneficial effects on the host, and their mechanism of action is through functions such as the production of bioactive compounds. In addition to dairy products, this bacterium has the potential to be isolated from non-dairy products and even plant products, and this issue can be an idea to isolate this bacterium from other sources and even identify new species with probiotic potential.
Keywords: Probiotic bacteria, Anti-microbial activity, Molecular identification
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Rahi A, Marhamatizadeh MH. Investigating the inhibitory effect of probiotic Lactobacillus species isolated from dairy products. Avicenna J Clin Microbiol Infect. 2022; 9(3):93-108. doi:10.34172/ajcmi.2022.3337
Introduction
The nutritional value and positive health effects of a dietary product in modern society are added advantages. The use of probiotics for dietary product development to manufacture functional foods with added advantages is well accepted (1-3). Probiotics are live microorganisms introduced in a specified manner and number into the dietary product for their ability to induce nutritional and health benefits to the consumer (4,5). The microorganisms used as probiotics are capable of improving digestion and absorption, preventing the growth of undesirable microorganisms in the colon and small intestine, reducing cholesterol, improving the immune system, and controlling allergic inflammation (6-8). The most common probiotics microorganisms are lactic acid bacteria (LAB) of the Lactobacillus genus (9).
LAB have been used for fermentation for ages and are still prominent in the food industry. LAB play an essential role in fermenting foods, extending the shelf life of fermented products, and imparting a beneficial influence on nutritional, health, and sensory characteristics of the end-products (10-12). These bacteria have no pathologic or toxic effect, can survive in the gastrointestinal tracts, and are tolerant to biliary salts (5,13). These properties make LAB the well-sorted probiotics for the food industry. Dairy products made from locally produced raw milk are still a highly important part of the daily diet with different inherent characteristics; however, fermentation by LAB is a common feature of all, making them a rich source for LAB screening, and traditional yogurts from different regions are a major source for the isolation of these probiotic microorganisms (10,14-17).
As part of normal flora, LAB are found in the human mouth, gut, and vagina (18). Members of this bacterial genus have the ability to produce organic acids such as lactic and acetic acids during the fermentation of carbohydrates. In addition, some of them can produce hydrogen peroxide and bacteriocin, which are anti-microbial compounds (19-22). Due to the production of such anti-microbial agents, LAB exhibit inhibitory effects on a variety of microorganisms. Various studies have confirmed the protective roles of LAB in intestinal and vaginal infections, as well as oral infections (19,23-25).
Yogurt is a traditional fermented milk product, popular in the rural areas of Iran. It is made of raw cow or sheep milk; the raw milk goes through spontaneous or starter fermentation to yield this traditional yogurt. Yogurt is a rich source for identifying new probiotic bacteria for industrial dairy use. Considering the fixed place of yogurt on the Iranian table and the highly useful and essential characteristics of the probiotic bacteria, in this study, probiotic LAB were isolated from yogurt samples, and their antimicrobial activity against a few pathogenic bacteria and fungi underwent evaluation. Because of their anti-microbial characteristics, these bacteria can be useful as bio-protective agents in controlling oral, gastrointestinal, and vaginal infections.
Materials and Methods
Sample Collection and Enrichment
Thirty yogurt samples were collected from different rural areas of Gachsaran, in the south of Iran. They were transferred to the laboratory in sterile containers and diluted to 10-7 using the Ringer solution (26). Then, 0.1 mL of different diluted suspensions were linearly inoculated on the agar MRS medium and incubated under micro-aerophilic conditions at 37°C for 48 hours (27). To separately enrich each cultured bacterium, a colony was transferred to the MRS broth medium and incubated at 37°C for 24-48 hours. To purify each microorganism, a loop of each cultured broth was linearly inoculated on the agar MRS medium and incubated under micro-aerophilic conditions at 37°C for 48 hours.
Phenotypic Identification of Isolates
Macroscopic and microscopic properties were used in the phenotypic identification of the isolates. Microscope and Gram-staining methods were used for microscopic identification. In addition, Gram-positive bacteria were selected and underwent biochemical tests such as catalase, oxidase, oxidative/fermentative, nitrate reduction, motility, triple sugar iron agar, and the ability to use sugars, including glucose, fructose, sucrose, lactose, galactose, trehalose, arabinose, xylose, starch, mannose, mannitol, rhamnosus, raffinose, cellobiose, and sorbitol (28).
Evaluation of Antimicrobial Activity Shown by the Isolates
The 48-hold cultures of lactobacilli were used to assess their antimicrobial activities. For this purpose, 10 mL of culture suspension of each isolate was separately harvested and centrifuged at 8500 rpm for 15 minutes. Next, 200 µm of the supernatant of each centrifuged medium was placed into the wells bored on Muller-Hilton agar plates cultured with E. coli, Staphylococcus aureus, Pseudomonas aeruginosa,and two fungi, namely, Aspergillus niger and Candida albicans. These plates were incubated at 35ºC for 24-48 hours, followed by measuring the diameter of the inhibition zone around each well. The size of the inhibition zone was considered as the potential of each isolate to produce bioactive compounds, and the anti-microbial activity of these compounds on the cultured bacteria and fungi was determined accordingly.
The Effect of Temperature and pH on the Growth and Antimicrobial Activity of Probiotic Isolates
To evaluate the effect of temperature and pH on the antimicrobial activity, each isolate was inoculated on MRS agar plates and incubated under micro-aerophilic conditions at 25, 35, 37, 40, and 45ºC for 72 hours (29). Furthermore, the pH of the culture media was adjusted to 2, 3, 4, 5, 6, 7, and 8 using HCl or 1M KOH. After inoculation, the isolates were incubated under the aforementioned condition at 37ºC for 24-48 hours. The growth rate of the isolates at different temperatures was measured by optical absorption, and their anti-bacterial properties were determined as described above.
Molecular Identification of Isolates by 16S rRNA Gene
DNA was extracted according to the Cardinal and Liz buffer method. The compounds used in the lysis buffer (Merck, Germany) included 1 M TRIS with a pH of 5.7, 5 M sodium chloride, 0.5 M EDTA, 2% SDS, and distilled water. Then, 0.8% agarose gel (Merck, Germany) was used on electrophoresis to check the quality of the extracted DNA. Specific primers (CinnaGen Company) were designed for the amplification of the 16S rRNA of Lactobacillus strains by Oligo5 software using 16S rRNA sequences present at NCBI (27F:AGAGTTTGATCMTGGCTCAG, 1492R: GGTTACCTTGTTACGACTT). The polymerase chain reaction (PCR) was performed by utilizing the following primers. The PCR was set up in a final volume of 25 μL using 12.5 μL of Mastermix (CinnaGen Company, Iran) with 0.4 μM of each primer (Forward and reverse) and 50 μL of the extracted DNA (30). Amplification was performed, including 5 cycles of initial denaturation (95°C, 4 minutes) and 30 cycles of denaturation (94°C, 1 minute), followed by annealing (57°C, 1 minute), extension (72°C, 2 minutes), and the final extension (72°C, 10 minutes).
Separation of Amplified DNA Fragments
PCR products were run on 0.8% agarose gel to separate the amplified DNA fragments. The amplified samples were mixed with loading buffer, loaded into the wells, and run. Following electrophoresis (VWR Scientific Company, model VWR105), gels were stained with ethidium bromide (Merck, Germany), and the amplified DNA bands of all the samples were observed and photographed by UV as single bands of approximately 1500 bp (Biometra model Germany BioDocAnalyze). PCR products were purified according to the instruction of the purification kite (Sigma-Aldrich, USA). Next, 50 µL of the purified DNA of each strain, along with both primers (a concentration of 50 pmol/µL) was sent for sequencing, which was performed by the Sanger sequencing method using the 3130XL Applied Biosystems-ABI sequencer.
Results
Results of Phenotypic Identification
Among the 30 collected samples, five samples growth on the MRS agar culture medium were considered as Lactobacillus species based on macroscopic observations such as colony appearance and colony color, as well as microscopic examinations such as gram-staining and cell morphology (Table 1).
Table 1.
Macroscopic and Microscopic Characteristics of Isolated Probiotic Bacteria
|
Sample
|
Microscopic Shape
|
Gram-staining
|
Colony-staining
|
Colony Appearance
|
Source
|
| X3 |
paired short rods |
Gram-positive with no spore |
Creamy |
Circular
mm 1 |
Yogurt |
| P |
Aggregated short rods |
Gram-positive with no spore |
Creamy white |
Ovoid
mm2-mm 5 |
Yogurt |
| PG |
Chained short rods |
Gram-positive with no spore |
Light white |
Circular
mm 1 |
Yogurt |
| X4 |
paired short rods |
Gram-positive with no spore |
White |
Circular
mm 1 |
Yogurt |
| A |
Short rods |
Gram-positive with no spore |
Creamy |
Circular
mm 1 |
Yogurt |
The phenotypic confirmation of Lactobacillus species was performed with biochemical tests such as catalase, oxidase, oxidative/fermentative, nitrate reduction, motility, and finally carbohydrate fermentation. The results were in accordance with Bergey’s Manual of Systematic Bacteriology (Table 2).
Table 2.
Biochemical Tests
|
Sample Test
|
A
|
PG
|
X4
|
P
|
X3
|
| Gram-positive reaction |
+ |
+ |
+ |
+ |
+ |
| Catalase activity |
- |
- |
- |
- |
- |
| Oxidase activity |
- |
- |
- |
- |
- |
| Oxidative/fermentative |
F |
F |
F |
F |
F |
| Production of CO2 from sugar |
- |
- |
- |
- |
- |
| Andol |
- |
- |
- |
- |
- |
| Citrate |
- |
- |
- |
- |
- |
| Motility |
- |
- |
- |
- |
- |
| Nitrate |
- |
- |
- |
- |
- |
| MR/VP |
- |
- |
- |
- |
- |
| Gelatin test |
- |
- |
- |
- |
- |
| Hemolysin test |
- |
- |
- |
- |
- |
| Arabinose |
+ |
- |
+ |
V |
V |
| Sculin |
+ |
V |
+ |
+ |
+ |
| Ribose |
+ |
+ |
+ |
+ |
+ |
| Glucose |
+ |
+ |
+ |
+ |
+ |
| Lactose |
V |
- |
+ |
+ |
+ |
| Manose |
+ |
+ |
- |
+ |
+ |
| Manitol |
+ |
+ |
V |
+ |
+ |
| Maltose |
+ |
+ |
- |
+ |
V |
| Rafinose |
- |
- |
- |
V |
+ |
| Sorbitol |
+ |
+ |
- |
- |
- |
| Sucrose |
V |
- |
- |
+ |
+ |
| Zylose |
+ |
+ |
+ |
+ |
+ |
| Melizitose |
V |
V |
V |
V |
+ |
| Trehalose |
- |
- |
+ |
+ |
+ |
Evaluation Results of Antimicrobial Activity
The results of the inhibitory effect of the bioactive compounds are listed in Table 3. It was found that these compounds had a growth inhibitory effect on the pathogens (Table 3). The most and least inhibitory effects were related to isolates A and X3, respectively, and S. aureus and A. niger were the most sensitive.
Table 3.
Evaluation of Bio-active Compounds Produced by Isolated Lactic Acid Bacteria and Their Spectrum
|
Isolates
|
Aspergillus
niger
|
Staphylococcus aureus
|
Salmonella Typhi
|
Escherichia coli
|
Candida albicans
|
| X3 |
12±1 |
10±1 |
15±1 |
13±1 |
14±1 |
| A |
14±1 |
17±1 |
16±1 |
17±1 |
15±1 |
| X4 |
13±1 |
16±1 |
15±1 |
14±1 |
12±1 |
| PG |
15±1 |
15±1 |
17±1 |
15±1 |
16±1 |
| P |
13±1 |
14±1 |
13±1 |
15±1 |
12±1 |
The Results of Determining the Optimal Temperature and pH for the Production of Bioactive Compounds
The results of the incubation of the isolates at different temperatures demonstrated that different isolates have the highest production of bioactive compounds at 37°C. The graph of the results is shown in Figure 1.
Based on the results of the incubation of the isolates at different pH rates, different isolates had the highest production of bioactive compounds at a pH rate of 7-8. Figure 2 depicts the graph of the results.
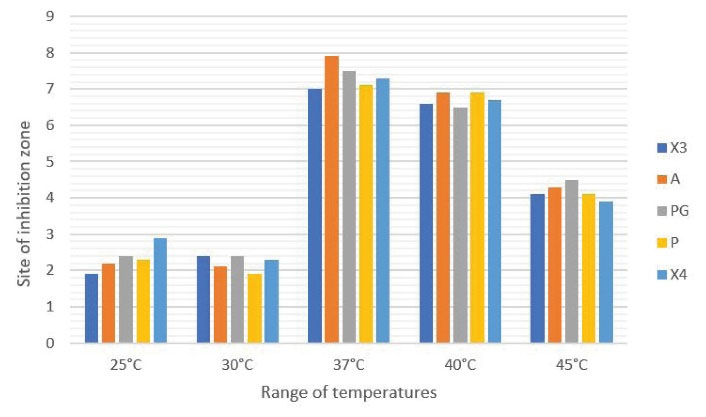
Figure 1.
The Results of the Production of Bioactive Compounds by Isolates Under Incubation at 25°C, 30°C, 37°C, 40°C, and 45°C.
.
The Results of the Production of Bioactive Compounds by Isolates Under Incubation at 25°C, 30°C, 37°C, 40°C, and 45°C.
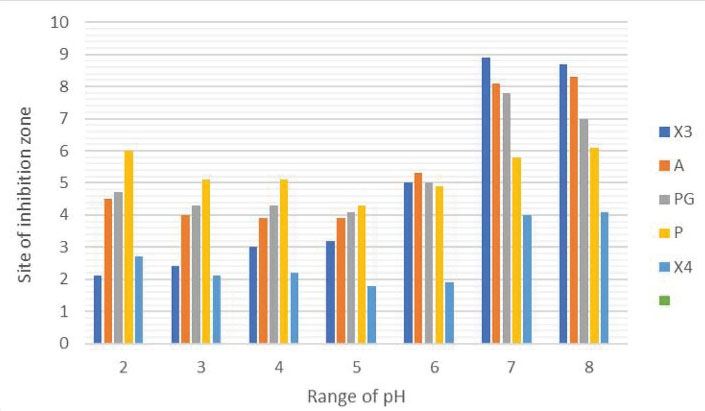
Figure 2.
The Results of the Production of Bioactive Compounds by Isolates Under Incubation at pH Rates of 2, 3, 4, 5, 6, 7, and 8.
.
The Results of the Production of Bioactive Compounds by Isolates Under Incubation at pH Rates of 2, 3, 4, 5, 6, 7, and 8.
Results of Molecular Identification
After sequencing, a comparison was made using the NCBI BLAST database, and X3 = Lactobacillus paracasei - 1171, X4 = Lactobacillus paracasei - 1171, and P = Lactobacillus plantarum strains were confirmed accordingly.
Discussion
According to the definition of the International Product Federation and the International Standard Organization, LAB are Gram-positive, non-spore-forming, catalase-negative, non-motile, nitrate reduction negative, and cytochrome oxidase negative, and unable to melt the gelatin and produce indole. All LAB have a fermentative metabolism and are strongly saccharolytic with lactic acid as the major final product of carbohydrate fermentation.
In this research, the most updated methods were used for the biochemical identification and molecular analysis of lactobacilli. The MRS medium was employed to isolate these bacteria in different studies, and then they were incubated at 37°C for 24 hours (29-31,32). This medium was also used in the study, and typical pure colonies of LAB were obtained accordingly. The method usually used in identifying Lactobacillus is biochemical analysis, including sugar fermentation. The number and type of fermented sugars observed in different studies can vary considerably. For example, Chammas et al applied the API kit, whereas other researchers investigated sugar fermentation without a kit (22,29,33,34). Another study considered the fermentation of 19 sugars, including amygdalin, galactose, cellobiose, lactose, mannitol, sucrose, melezitose, melibiose, raffinose, rhamnosus, sorbitol, Torre trehalose, maltose, ribose, and gentiobiose for the biochemical identification of LAB (22). By comparing biochemical identification results with the tables presented in Bergy’s manual, different strains were identified, including Lactobacillus casei, Lactobacillus plantarum, Lactobacillus bifidum, Lactobacillus reuteri, and rhamnosus. However, physiological and biochemical tests have some limitations, and there are physiological similarities among a number of isolates. In addition, the results of these tests are not always accurate, indicating the details of microbial diversity in fermented milk products. Hence, molecular methods have been recommended to confirm the phenotypic identification of the isolated strains. In the case of the accurate identification of Lactobacillus strains by molecular markers, a number of methods have been developed, including RAPD (33) DNA-DNA hybridization, as well as the comparison of 16S rRNA sequences (31,32). It has been noted that the last two methods are more accurate. Considering that the fermented sugars by many isolated strains did not match any specific strains in Bergy’s manual, it was impossible to identify them phenotypically, thus the sequence of their 16S rRNAs was determined and compared with the sequences of other reported bacteria using the NCBI database, leading to their identification.
Samples containing two strains of X3 and X4 were collected at remote destinations. The environmental conditions affecting the growth of these organisms were completely different, thus their biochemical properties (e.g., sugar fermentation) were unique. However, their close genetic relationship was highly interesting. The differences observed in these bacteria related to the consumption of sugar and lack of compliance with the Bergy’s manual might be the result of one or more mutations in genes related to intermediary enzymes involved in sugar fermentation. Perhaps, during evolution and in response to environmental conditions, they had no need to ferment these sugars, and the gene(s) was/were mutated or deleted. It appears that the environmental stability of these bacteria resulting from the beliefs of individuals or families has kept the strain(s) of one family away from the others, and thus these strains had no need to remedy or reform their biochemical pathways.
Probiotic bacteria through different mechanisms, including pH reduction, competition for cell receptors, and the production of anti-microbial compounds (e.g., bacteriocins, Lactic and acetic acids, hydrogen peroxide, acetaldehyde, and ammonia) can affect bacterial metabolism or the toxin production and show inhibitory effects on many microorganisms. Lactobacilli isolated from dairy products are capable of inhibiting S. aureus, E. coli, and Bacillus (35). Likewise, the findings of this research indicated that Lactobacilli isolated from yogurt have inhibitory effects on five strains, and the maximum sensitivity was related to S. aureus and Aspergillus (Table 3). Heydari et al, isolated 17 strains of lactic acid bacteria from dairy products and tested them for their anti-microbial activity, They reported the largest inhibition zone (20 mm diameter) for Staphylococcu aureus and the minimum inhibition zone (1mm diameter) for Entrobacter aeruginosa (36). Similarly, in this study, the largest inhibition zone (17 mm in diameter) belonged to S. aureus (Table 3). Morever Soltani et al found that Lactobacillus spp. isolates is able to inhibit the overall growth of E. coli (2.10 mm) and Staphylococcus aureus(8.9 mm) (37). According to the findings of this research and the results of similar research (38), the anti-microbial activity of LAB against Gram-positive bacteria is greater against Gram-negative ones.
Conclusion
Different lactic acid probiotic bacteria were isolated and identified from traditional yogurt samples collected from around Gachsaran. Among the isolates that were sent for molecular identification, X3 and X4 were isolated from completely different places. However, they were completely similar from a molecular point of view, representing the spatial distribution of probiotic bacteria that are genetically similar in different places. Probiotic bacteria have extremely beneficial effects on the host, and their mechanism of action is through functions such as the production of bioactive compounds. In addition to dairy products, this bacterium can be isolated from non-dairy products and even plant products; accordingly, this issue can be an idea to isolate this bacterium from other sources and even identify new species with probiotic potential.
Acknowledgements
The authors would like to thank Dr. Hossein Fattahi for his imperative support in laboratory examinations.
Authors’ Contribution
MHM and AR conceived the idea. AR did the work and wrote the manuscript. MHM reviewed the manuscript. AR performed the statistical analysis.
Conflict of Interests
The authors declare that there is no conflict of interests.
Ethical Approval
The contemporary survey was accepted by the Ethics Research Committee of the Department of Food Hygiene, Veterinary Faculty, Kazerun Branch, Islamic Azad University, Kazerun, Iran (with number 93/1905 on December 22, 2014).
References
- Granato D, Branco GF, Nazzaro F, Cruz AG, Faria JAF. Functional foods and nondairy probiotic food development: trends, concepts, and products. Compr Rev Food Sci Food Saf 2010; 9(3):292-302. doi: 10.1111/j.1541-4337.2010.00110.x [Crossref] [ Google Scholar]
- Hasler CM. Functional foods: benefits, concerns and challenges-a position paper from the American Council on Science and Health. J Nutr 2002; 132(12):3772-81. doi: 10.1093/jn/132.12.3772 [Crossref] [ Google Scholar]
- Siró I, Kápolna E, Kápolna B, Lugasi A. Functional food Product development, marketing and consumer acceptance--a review. Appetite 2008; 51(3):456-67. doi: 10.1016/j.appet.2008.05.060 [Crossref] [ Google Scholar]
- Guarner F, Schaafsma GJ. Probiotics. Int J Food Microbiol 1998; 39(3):237-8. doi: 10.1016/s0168-1605(97)00136-0 [Crossref] [ Google Scholar]
- Kailasapathy K, Chin J. Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol Cell Biol 2000; 78(1):80-8. doi: 10.1046/j.1440-1711.2000.00886.x [Crossref] [ Google Scholar]
- Grashorn M, editor Use of probiotics, digestion enhancers and growth promoters in diets for heavy turkeys. Proceedings of 10th European Poultry Conference; 1998; Jerusalem.
- Tomaro-Duchesneau C, Jones ML, Shah D, Jain P, Saha S, Prakash S. Cholesterol assimilation by Lactobacillus probiotic bacteria: an in vitro investigation. Biomed Res Int 2014; 2014:380316. doi: 10.1155/2014/380316 [Crossref] [ Google Scholar]
- Ashraf R, Shah NP. Immune system stimulation by probiotic microorganisms. Crit Rev Food Sci Nutr 2014; 54(7):938-56. doi: 10.1080/10408398.2011.619671 [Crossref] [ Google Scholar]
- Fijan S. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health 2014; 11(5):4745-67. doi: 10.3390/ijerph110504745 [Crossref] [ Google Scholar]
- Abd El Gawad IA, Abd El Fatah AM, Al Rubayyi KA. Identification and characterization of dominant lactic acid bacteria isolated from traditional Rayeb milk in Egypt. J Am Sci 2010; 6(10):728-35. [ Google Scholar]
- Olaoye OA, Onilude AA, Dodd CE. Identification of Pediococcus spp from beef and evaluation of their lactic acid production in varying concentrations of different carbon sources. Adv Nat Appl Sci 2008; 2(3):197-207. [ Google Scholar]
- De Vuyst L, Leroy F. Bacteriocins from lactic acid bacteria: production, purification, and food applications. J Mol Microbiol Biotechnol 2007; 13(4):194-9. doi: 10.1159/000104752 [Crossref] [ Google Scholar]
- Hasani M, Hesari J, Farajnia S, Moghaddam Vahed M. Technological characterisation of predominant lactobacilli isolated from traditional Lighvan cheese. J Food Res 2011; 21(4):539-51. [ Google Scholar]
- Ambadoyiannis G, Hatzikamari M, Litopoulou-Tzanetaki E, Tzanetakis N. Probiotic and technological properties of enterococci isolates from infants and cheese. Food Biotechnol 2004; 18(3):307-25. doi: 10.1081/FBT-200035024 [Crossref] [ Google Scholar]
- Sharifi Yazdi MK, Davoodabadi A, Khesht Zarin HR, Tajabadi Ebrahimi M, Soltan Dallal MM. Characterisation and probiotic potential of lactic acid bacteria isolated from Iranian traditional yogurts. Italian Journal of Animal Science 2017; 16(2):185-8. doi: 10.1080/1828051X.2016.1222888 [Crossref] [ Google Scholar]
- Nishimura M, Ohkawara T, Tetsuka K, Kawasaki Y, Nakagawa R, Satoh H. Effects of yogurt containing Lactobacillus plantarum HOKKAIDO on immune function and stress markers. J Tradit Complement Med 2016; 6(3):275-80. doi: 10.1016/j.jtcme.2015.07.003 [Crossref] [ Google Scholar]
- Simova ED, Beshkova DB, Dimitrov ZP. Characterization and antimicrobial spectrum of bacteriocins produced by lactic acid bacteria isolated from traditional Bulgarian dairy products. J Appl Microbiol 2009; 106(2):692-701. doi: 10.1111/j.1365-2672.2008.04052.x [Crossref] [ Google Scholar]
- Ahrné S, Nobaek S, Jeppsson B, Adlerberth I, Wold AE, Molin G. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J Appl Microbiol 1998; 85(1):88-94. doi: 10.1046/j.1365-2672.1998.00480.x [Crossref] [ Google Scholar]
- McGroarty JA, Tomeczek L, Pond DG, Reid G, Bruce AW. Hydrogen peroxide production by Lactobacillus species: correlation with susceptibility to the spermicidal compound nonoxynol-9. J Infect Dis 1992; 165(6):1142-4. doi: 10.1093/infdis/165.6.1142 [Crossref] [ Google Scholar]
- Kanatani K, Oshimura M, Sano K. Isolation and characterization of acidocin A and cloning of the bacteriocin gene from Lactobacillus acidophilus. Appl Environ Microbiol 1995; 61(3):1061-7. doi: 10.1128/aem.61.3.1061-1067.1995 [Crossref] [ Google Scholar]
- Green G, Dicks LM, Bruggeman G, Vandamme EJ, Chikindas ML. Pediocin PD-1, a bactericidal antimicrobial peptide from Pediococcus damnosus NCFB 1832. J Appl Microbiol 1997; 83(1):127-32. doi: 10.1046/j.1365-2672.1997.00241.x [Crossref] [ Google Scholar]
- Van der Meulen R, Scheirlinck I, Van Schoor A, Huys G, Vancanneyt M, Vandamme P. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl Environ Microbiol 2007; 73(15):4741-50. doi: 10.1128/aem.00315-07 [Crossref] [ Google Scholar]
- Tannock GW. The microecology of lactobacilli inhabiting the gastrointestinal tract. In: Marshall KC, ed. Advances in Microbial Ecology. Boston, MA: Springer; 1990. p. 147-71. 10.1007/978-1-4684-7612-5_4.
- Andreu A, Stapleton AE, Fennell CL, Hillier SL, Stamm WE. Hemagglutination, adherence, and surface properties of vaginal Lactobacillus species. J Infect Dis 1995; 171(5):1237-43. doi: 10.1093/infdis/171.5.1237 [Crossref] [ Google Scholar]
- Arihara K, Ogihara S, Mukai T, Itoh M, Kondo Y. Salivacin 140, a novel bacteriocin from Lactobacillus salivarius subsp salicinius T140 active against pathogenic bacteria. Lett Appl Microbiol 1996; 22(6):420-4. doi: 10.1111/j.1472-765x.1996.tb01194.x [Crossref] [ Google Scholar]
- Hasan RN, Ali MR, Shakier SM, Khudhair AM, Hussin MS, Kadum YA. Antibacterial activity of aqueous and alcoholic extracts of Capsella bursa against selected pathogenic bacteria. Am J BioSci 2013; 1(1):6-10. doi: 10.11648/j.ajbio.20130101.12 [Crossref] [ Google Scholar]
- Goodfellow M, Kämpfer P, Busse HJ, Trujillo ME, Suzuki KI, Ludwig W, et al. Bergey’s Manual of Systematic Bacteriology: Volume 5: The Actinobacteria. Springer Science & Business Media; 2012.
- Kandler O, Nobert W. Bergeys Mannual of Systematic. New York: Springer; 1989.
- Pourahmad R, Mazaheri-Assadi M, Mirdamadi S. Isolation and identification of Iranian native yoghurt starters. Pajouhesh & Sazandegi 2004; 17(4):42-8. [ Google Scholar]
- Tamarapu S. Development of a multiplex PCR for detection of Staphylococcus aureus in skim milk and cheddar cheese: Mississippi State University; 2000.
- Watanabe K, Fujimoto J, Sasamoto M, Dugersuren J, Tumursuh T, Demberel S. Diversity of lactic acid bacteria and yeasts in Airag and Tarag, traditional fermented milk products of Mongolia. World J Microbiol Biotechnol 2008; 24(8):1313-25. doi: 10.1007/s11274-007-9604-3 [Crossref] [ Google Scholar]
- Chammas GI, Saliba R, Corrieu G, Béal C. Characterisation of lactic acid bacteria isolated from fermented milk “laban”. Int J Food Microbiol 2006; 110(1):52-61. doi: 10.1016/j.ijfoodmicro.2006.01.043 [Crossref] [ Google Scholar]
- Fitzsimons NA, Cogan TM, Condon S, Beresford T. Spatial and temporal distribution of non-starter lactic acid bacteria in Cheddar cheese. J Appl Microbiol 2001; 90(4):600-8. doi: 10.1046/j.1365-2672.2001.01285.x [Crossref] [ Google Scholar]
- Ayhan K, Durlu-Özkaya F, Tunail N. Commercially important characteristics of Turkish origin domestic strains of Streptococcus thermophilus and Lactobacillus delbrueckii ssp bulgaricus. Int J Dairy Technol 2005; 58(3):150-7. doi: 10.1111/j.1471-0307.2005.00206.x [Crossref] [ Google Scholar]
- Nowroozi J, Mirzaii M, Norouzi M. Study of Lactobacillus as probiotic bacteria. Iran J Public Health 2004; 33(2):1-7. [ Google Scholar]
- Heidari Z, Ghasemi MF, Modiri L. Antimicrobial activity of bacteriocin produced by a new Latilactobacillus curvatus spLAB-3H isolated from traditional yogurt. Arch Microbiol 2021; 204(1):101. doi: 10.1007/s00203-021-02641-8 [Crossref] [ Google Scholar]
- Soltani N, Abbasi S, Baghaeifar S, et al. Antibacterial and antibiofilm activity of Lactobacillus strains secretome and extraction against Escherichia coli isolated from urinary tract infection. Biotechnol Rep (Amst) 2022;36:e00760. Published 2022 27. 10.1016/j.btre.2022.e00760.
- Ouwehand AC, Salminen SJ. The health effects of cultured milk products with viable and non-viable bacteria. Int Dairy J 1998; 8(9):749-58. doi: 10.1016/s0958-6946(98)00114-9 [Crossref] [ Google Scholar]