Avicenna Journal of Clinical Microbiology and Infection. 8(1):17-22.
doi: 10.34172/ajcmi.2021.04
Original Article
Contamination of Bovine Milk With Brucella spp.: A Current Public Health Menace in Kurdistan Province of Iran
Mohammad Hadi Asgari 1  , Elham Ahmadi 2, *
, Elham Ahmadi 2, * 
Author information:
1Graduated from Faculty of Veterinary Medicine, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran
2Department of Pathobiology, Sanandaj Branch, Islamic Azad University, Sanandaj, Iran
Abstract
Background: Brucellosis, a deteriorating zoonotic disease, is very common in most parts of Iran. Consumption of contaminated milk and dairy products is one of the most significant ways for transmission of the infections to human. Since the close rearing of cattle and sheep is practiced in Kurdistan province of Iran, the infection of cow with non-specific species is not out of mind. The present study aimed to determine the frequency of bovine milk contamination with zoonotic Brucella spp.
Methods: A total of 240 milk samples, equally from traditional and industrialized dairy farms, were collected aseptically. Conventional microbiological method was used for isolation of the bacterium, followed by the genotypic identification of the isolates. Moreover, direct molecular processing of the samples was carried out for detection of the bacterial genome. The positive samples were further genotypically assessed to identify the contamination as Brucella abortus or Brucella melitensis.
Results: In general, 16 (6.66%) and 15 (6.25%) of the samples were contaminated with Brucella spp. in phenotypic and genotypic methods, respectively. The proportion of contamination with B. abortus and B. melitensis in phenotypic and genotypic methods were 5% and 1.66%, and 5% and 1.25%, respectively. The overall rate of contamination in traditional milk samples was more than industrialized samples.
Conclusions: Contamination of bovine milk with Brucella spp. is a serious threat to public health in the studied region. Continuous vaccination, application of test and slaughter policy, and presumption of pasteurized milk and dairy products are highly recommended.
Keywords: Bovine milk, Brucella abortus, Brucella melitensis, Western Iran
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Brucella spp., the etiological agent of brucellosis in domestic animals and Malta fever in human, is a Gram negative, facultative intracellular aerobic microorganism (1). The World Health Organization (WHO) has categorized the disease as one of the seven neglected zoonotic infections in human despite its worsening implications (2). The genus Brucella comprises of 12 species based on the main host and pathogen (3). Among the various species, Brucella abortus, Brucella suis, and Brucella melitensis cause infections in human, where the latter species is considered the most infectious one (4). Following the infection of a female animal, the bacterium affects the reproductive system, and finally localizes in mammary gland and retromammary lymph nodes everlastingly. Continues shedding of the bacteria in milk imposes a public health hazard (5). The main routes for bacterial transmission to human is through not only consumption of unpasteurized milk and dairy products, but also close contact with infected animals, as well as inhalation (6).
The Middle East and Central Asia are the hotspots with the highest incidence of the infection among livestock and human (7). In Iran, vaccination with S19 or RB51 strains in cattle and Rev1 strain in sheep/goat, as well as test/eradication policy in domestic animals have been launched to control the infection in dairy industry (8,9). Although serological assays including milk ring test (MRT), Rose Bengal slide agglutination, and standard tube agglutination (STA) tests are frequently used in brucellosis surveillance and control programs, they may produce false-negative (10,11) or false-positive (12) results. Moreover, the lack of knowledge regarding the Brucella species involved in a brucellosis case in the serological tests is another negative point of this method. As a consequence, the probability of ascertaining the infection source and, therefore, applying the proper control procedures may have been precluded (13). In comparison, culture and microbiological isolation, as the gold standard technique, as well as molecular methods for detectionof Brucella spp. may be more informative and trustable (10).
Although Brucella spp. attack preferred hosts, some species may infect the non-preferred hosts in close rearing system of different animal species. Due to the scant of recent information regarding the prevalence of Brucella in bovine milk and keeping cows in close proximity of small ruminants in the same barn, the present study was conducted to evaluate the frequency of B. abortus and B. melitensis in cow milk samples collected from Kurdistan province of Iran.
Methods
Sample Collection
In a cross-sectional study, from December 2018 to May 2019, a total of 240 milk samples, equally from industrialized and agrarian farms in all over Kurdistan province of Iran, were collected aseptically in sterile bottle glasses after disinfecting four teats. The samples were chilled until their delivery to the laboratory within maximum five hours and divided into two parts. One part was scrutinized for bacteriological isolation and the other one was inactivated at 65°C for 30 min, and stored at -20°C until molecular analysis (14).
Bacterial Culture
Following the centrifuging of the milk samples in 3000 g for 15 minutes, the pellet and cream were streaked on Brucella selective agar (Quelab, Canada) complemented with Brucella selective supplement (Oxoid, UK). The plates were incubated at 37°C for 14 days under 5%-10% CO2 tension and examined for Brucella spp. on daily base after the fourth day (15). Resultant colonies representing the morphology of Brucella spp. were further subcultured on Blood agar (BA, Quelab, Canada) containing 7% defibrinated sheep blood sample. Conventional identification of the isolates as Brucella spp. was based on Gram and modified Ziehl-Neelsen (MZN) stainings, catalase, oxidase, H2S production, and nitrate and urea reactions (16).
Genomic DNA Extraction
For initiation, the samples were thawed, 1.5 mL of each one was poured into sterile two μL tube, and centrifuged for 10 min at 6000 g. Three layers including the cream (the upper fat), milk whey, and the protein deposition (the lower layer) were separated. The milk whey was collected and discarded, then 200 μL Tris-EDTA (TE) solution was added to each microtube and homogenized thoroughly (8). In addition, the overnight culture of the bacteria isolated in phenotypic method was used for DNA extraction.
DNA was extracted from both pure culture of the isolates and directly from each milk sample using Gram Negative Bacterial DNA Extraction Kit (CinnaGen, Iran, Cat no. EX6011), in accordance with the manufacturer’s guidelines.
Molecular Assessment
Molecular detection of Brucella spp. was carried out with the partial amplification of bcsp31 gene using primer pair and thermal condition introduced elsewhere (17) (Table 1). In the next round of polymerase chain reaction (PCR) reaction, the identified bacteria were categorized as B. abortus (all biovars) or B. melitensis (all biovars) based on the method represented by Whatmore et al (18) and Bricker and Halling (19) (Table 1). All molecular assays were repeated twice.
Table 1.
Details of Primer Sequences and Thermal Conditions Used in the Present Study
|
Gene
|
Primer Sequence (5ʹ→3ʹ)
|
PCR Thermal Condition
|
Product Size (bp)
|
Reference
|
|
Initial Denaturation
|
Denaturation
|
Annealing
|
Extension
|
Final Extension
|
Bcsp31
(Brucella spp.)
|
TGGCTCGGTTGCCAATATCAA
CGCGCTTGCCTTTCAGGTCTG
|
95°C for 4 min |
94°C for 1 min |
60°C for 1 min |
72°C for 45 sec |
72°C for 5 min |
223 |
(17)
|
| 40 cycles |
|
Omp25 (B. abortus)
|
ATGCGCACTCTTAAGTCTC
GCCSAGGATGTTGTCCGT
|
95 °C for 4 min |
94 °C for 1 min |
59 °C for 1 min |
72 °C for 1 min |
72 °C for 7 min |
490 |
(18)
|
| 35 cycles |
|
IS711 (B. melitensis)
|
AAATCGCGTCCTTGCTGGTCTGA
TGCCGATCACTTAAGGGCCTTCAT
|
95 °C for 4 min |
95 °C for 1:15 min |
55/5 °C for 2 min |
72 °C for 1 min |
72 °C for 7 min |
731 |
(19)
|
| 30 cycles |
RB51 and Rev1 strains (kindly provided by Veterinary Administration Office of Kurdistan province) were used as positive controls and sterile distilled water was exercised as negative control in the survey. The amplicons were electrophoresed into 1.2% agarose gel in 80 V for 70 minutes and visualized under UV light.
Statistical Analysis
The statistical association of the frequency of Brucella spp. and the two studied species in phenotypic and genotypic methods with the rearing system (traditional and industrialize) was analyzed in SPSS software (version 21.0, Chicago, IL) using chi-square test. A P value ≤ 0.05 was considered statistically significant.
Results
Cultivation of the milk samples demonstrated the growth of pinpoint, dew-drop typical round, glistening, convex, and translucent colonies in 16 (6.66%) cases. An individual colony from each plate was subcultured on BA. The pure colonies with an appearance of pink small cocobacilli in Gram and MZN stainings, catalase and oxidase positive, representing positive reactions in nitrate reduction and urease production tests, were biochemically identified as Brucella spp. In addition, a trace of H2S production in four (1.66%) samples and no evidence of H2S production in 12 (5%) samples primarily categorized the isolates as B. melitensis and B. abortus, respectively. Thirteen (5.41%) and three (1.25%) isolates were obtained from traditional and industrialized milk samples. The frequency of the isolates in traditional and industrialized milk samples are illustrated in Table 2. All of the isolates were molecularly confirmed as Brucella spp. in the bcsp31 (genus)-specific PCR reaction (Figure 1). In the next step, the isolates generated the expected amplicons in species-specific reactions, confirming as B. abortus and B. melitensis (Figures 2 and 3)
Table 2.
Details Regarding the Frequency of Brucella Spp. in Milk Samples
|
Samples
|
No. (%) of Samples
|
No. (%) of
Brucella
spp. in Phenotypic Analysis
|
No. (%) of
B. abortus
in Phenotypic Analysis
|
No. (%) of
B.
melitensis
in Phenotypic Analysis
|
No. (%) of
Brucella
spp. in Molecular Analysis
|
No. (%) of
B. abortus
in Molecular Analysis
|
No. (%) of
B.
melitensis
in Molecular Analysis
|
| Traditional milk samples |
120 (50%) |
13 (5.41%) |
10 (4.16%) |
3 (1.25%) |
12 (5%) |
10 (4.16%) |
2 (0.83%) |
| Industrialized milk samples |
120 (50%) |
3 (1.25%) |
2 (0.83%) |
1 (0.41%) |
3 (1.25%) |
2 (0.83%) |
1 (0.41%) |
| Total |
240 (100%) |
16 (6.66%) |
12 (5%) |
4 (1.66%) |
15 (6.25%) |
12 (5%) |
3 (1.25%) |
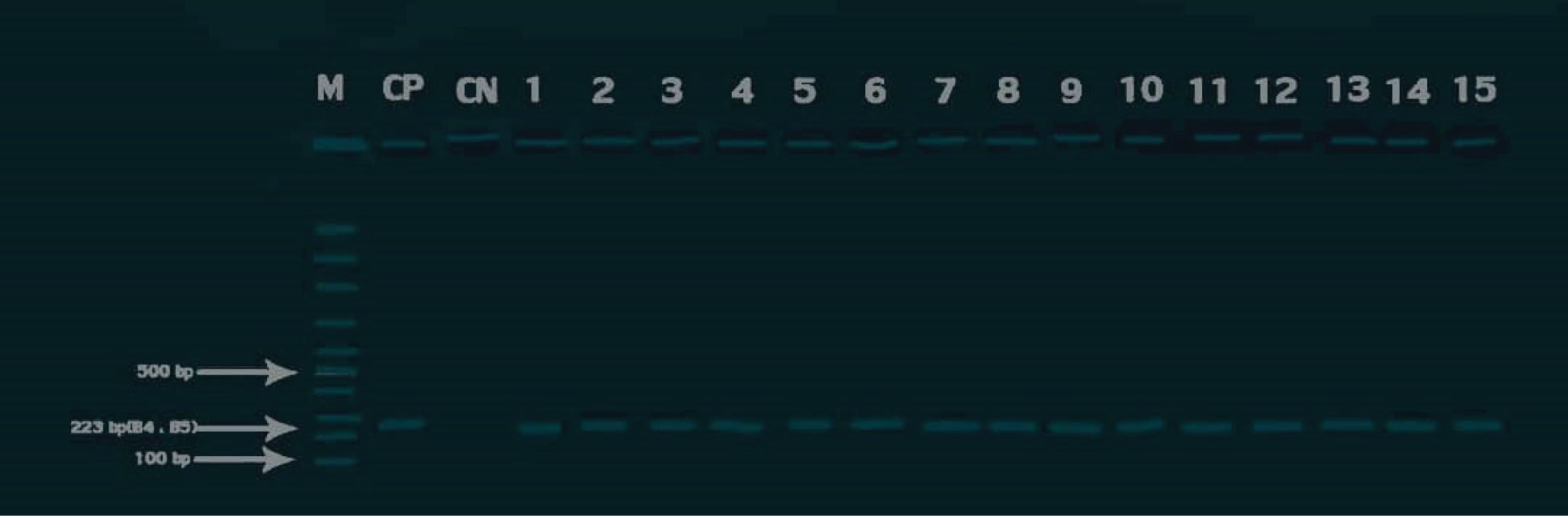
Figure 1.
Agarose Gel Electrophoresis of PCR Products Generated From Bcsp31 gene Amplification in Brucella spp. M: 100 bp DNA Ladder (Sinaclon, Iran), CP: Positive Control (RB51 Strain), CN: Negative Control, Lanes1-15: Field Samples.
.
Agarose Gel Electrophoresis of PCR Products Generated From Bcsp31 gene Amplification in Brucella spp. M: 100 bp DNA Ladder (Sinaclon, Iran), CP: Positive Control (RB51 Strain), CN: Negative Control, Lanes1-15: Field Samples.
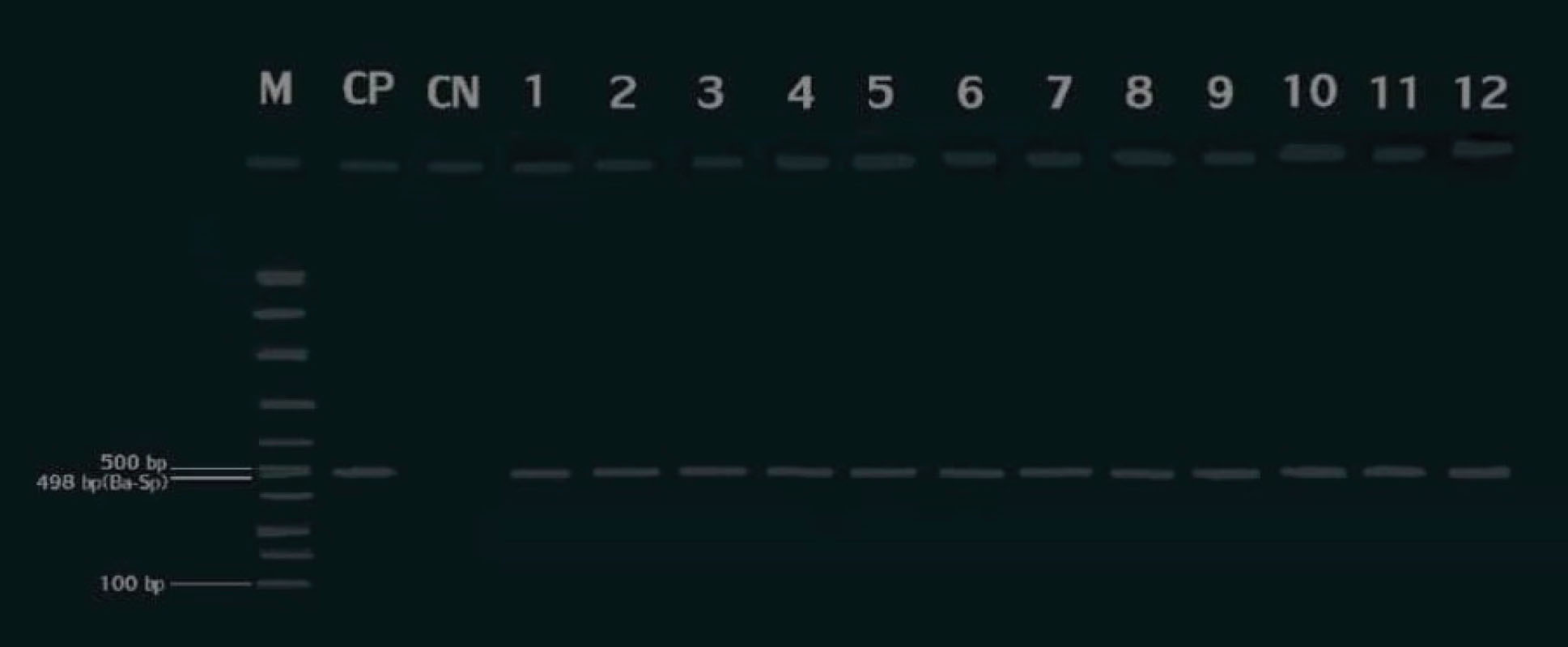
Figure 2.
Agarose Gel Electrophoresis of PCR Products Generated From Omp25 gene Amplification in B. abortus. M: 100 bp DNA Ladder (Sinaclon, Iran), CP: Positive Control (RB51 Strain), CN: Negative Control, Lanes1-12: Field Samples.
.
Agarose Gel Electrophoresis of PCR Products Generated From Omp25 gene Amplification in B. abortus. M: 100 bp DNA Ladder (Sinaclon, Iran), CP: Positive Control (RB51 Strain), CN: Negative Control, Lanes1-12: Field Samples.

Figure 3.
Agarose Gel Electrophoresis of PCR Products Generated from IS711 gene Amplification in B. Melitensis. M: 100 Bp DNA Ladder (Sinaclon, Iran), CP: Positive Control (Rev1 Strain), CN: Negative Control, Lanes1-3: Field Samples.
.
Agarose Gel Electrophoresis of PCR Products Generated from IS711 gene Amplification in B. Melitensis. M: 100 Bp DNA Ladder (Sinaclon, Iran), CP: Positive Control (Rev1 Strain), CN: Negative Control, Lanes1-3: Field Samples.
Moreover, the contamination of 15 (6.25%) out of 240 milk samples with Brucella spp. was detected in molecular approach (Figure 1), among which 12 (5%) and three (1.25%) were recognized as B. abortus and B. melitensis(Figures 2 and 3), respectively. The details of Brucella spp. contamination in traditional and industrialized milk samples are depicted in Table 2.
Besides, the statistical relationships were observed between the frequencies of Brucella spp. (P ≤ 0.05) and B. abortus (P ≤ 0.05) with the rearing system, which was the opposite to the result obtained for B. melitensis (P > 0.05).
Discussion
Milk and dairy products are incriminated as potential vectors for transmission of some notorious zoonotic diseases to human. In some previous studies undertaken by the author and colleagues in the studied region, the contamination of milk with Coxiella burnetii and aflatoxin M1 (20), Shige-toxigenic Escherichia coli (21), and methicillin resistant Staphylococcus aureus has also been proved (22). Based on the mistaken beliefs regarding the benefits of unpasteurized milk and dairy products compared to pasteurized form, the consumption of these products are very popular in the district. The results represented the average 6% frequency of milk contamination with Brucella spp., which is a matter of concern due to the worsening implications of the disease in both veterinary and public sector. On the other hand, empirical evidence has manifested the habit of using communal grazing or keeping different species of animals (cattle, sheep, and goat) tethered or at limited pastures. This provides favorable conditions for propagation of non-preferred bacterial species among domestic animals (23). The spillover of B. melitensis from sheep and goat to cattle has previously been demonstrated in other studies (8,14). Generally, the animal breeding capacity of Kurdistan, an interest in consumption of unpasteurized milk and dairy products, and deregulation of trade and decreased animal traffic control in borders with neighboring countries are the plausible reasons for the burden of the infection in the given province (8,24-26). Notably, the results implied that the infection in small ruminantsʹ population is currently prevalent in the region which should be considered in the future epizootiological surveillance programs.
Despite the application of routine vaccination and eradication programs for Brucella spp. in Iran, the serology screening is not trustable due to the positive and negative false results (10-12). Sampling at an early stage of the infection (e.g., within the first 14 days (27), reduced antibody titers over time (28), generation of a shorter duration of humoral antibody response in cattle due to infection with B. suis (29), previous vaccination against brucellosis (30), and latent infectious status in the cases of inter-uterus infection or in the early postnatal period (5) are possible reasons producing false negative results. In addition, false positive serological results might be generated because of cross-reaction with E. coli, Vibrio cholera, Yersinia enterocolitica, and Francisella tularensis (5).
Although the bacterial isolation is the gold standard diagnostic method for Brucella spp. (31), there are some obstacles. A plausible reason for avoiding the application of phenotypic method in a routine manner may be related to the highly hazardous and contagious zoonotic nature of the bacterium, which requires appropriate biosecurity facilities and personnel having expertise in diagnostic laboratory (32). Besides, the bacterium is classified as a class-B bio-weapon and the procedure is time-consuming (32). In our study, a total number of 16 Brucella spp. was isolated using microbiological procedure. All of the isolates were molecularly confirmed as the genus and further characterized as B. abortus and B. melitensis. Some other internal (33,34) and external (35,36) researches have also been documented the identification of zoonotic Brucella spp. in ruminants’ milk samples using conventional bacteriological method and molecular confirmation of the isolates using PCR. The reported contamination rate was from 1.28% to 25% (33-36).
On the other hand, not only does the replacement of a fast, simple, and accurate complementary diagnostic method for direct molecular detection of the bacterium in milk provide precise detection of the infection, but it also protects laboratory staff against contamination. Herein the subtle higher identification rate of the bacterium in the phenotypic method compared to the direct molecular approach may be explained by the fact that the different compounds of the milk such as proteins and fats may exert inhibitory effect on direct DNA extraction protocols. Also, a drawback of the used kit is that it was not specific for bacterial DNA extraction from milk samples. Consequently, this may influence the final outcome. Remarkably, some factors, namely the amount of the bacteria shedding through the milk, disease phase, and fastidiously-growing nature of the genus may affect the bacterial detection ratio (37). Similar to our study, another survey compared the culture and PCR methods for detection of Brucella spp. in milk samples (38). The results represented a significant higher proportion of bacterial detection in PCR in comparison with the phenotypic method, which is in contrast with the data obtained herein. A possible reason for this incompatibility may be related to the DNA extraction method applied in the both studies.
It is assumed that the frequency of brucellosis is underestimated in the present research as no ovine and caprine milk samples were evaluated. Likewise, the predominant biovar of B. abortus identified in different internal studies in Iran is biovars 3 (8,39-41). RB51 or S19 strains of the bacterium, used as a vaccine, are biovar 1 strains. The latter may be also disposed in milk like a wild-type strain and inaccurately recognized as a severe strain (8). Therefore, further research is mandatory to draw firm conclusions about the exact distribution of wild-type Brucella spp. in milk. Some PCR-based techniques can simply discriminate between the wild-type and vaccine strains based on the amplicon size (32,42).
Considering the endemic status of brucellosis in Iran, epidemiological investigation of the disease is of great significance. Some literature has documented the frequency of Malta fever or brucellosis in recent years in Kurdistan province of Iran. Demographic information relating to the distribution of Malta fever among 1997 to 2003 documented the statistical association of gender and place of living with the frequency of the disease, as males in villages and bricklayer in cities were the most affected groups. Moreover, the highest level of incidence was in 2003 and the lowest in 2000 (43). Norouzinezhad et al have assessed the epidemiological characteristics and trend of the incidence of human brucellosis in Kurdistan province from 2009 to 2016. The results depicted the highest incidence as 103.54 in 100 000 in 2014 and the lowest as 23.86 in 100 000 in 2010. According to an 8-year analysis, the highest incidence rate was seen in Bijar county among farmers, housewives, ranchers, male sex, rural dwellers, and those aged 24-25 years. Majority of the patients reported contact with livestock (26). In another study analysing the seroepidemiology of brucellosis in 2014, the rate was 6.4% with the highest burden among butchers (12%) (24). Besides, the distribution of Brucella spp. in bovine and caprine milk in 2012 in Kurdistan was stated to have been 33.33% and 44% with the frequency of B. abortus and B. melitensis as 15% and 3.33%, and 2% and 30%, respectively (8). The frequency of the infection in cow milk and its traditional products in Isfahan and Chaharmahal and Bakhtiari provinces of Iran in 2012 was reported to have been 1% B. abortus in milk, 2.5% B. abortus and B. melitensis in cheese, and 1% B. abortus in cream (44). In contrast, the all 12 Brucella spp. detected in raw and unpasteurized bulk cow milk tanks of traditional domestic dairy sale centers in Khorramabad was B. abortus (9). The plausible explanations for the discrepancy in various studies may be related to the type and number of sampling, methodology, geographical and socioeconomical conditions, vaccination, and disease controlling measures (9,45). Moreover, the frequency of Brucella spp. DNA in bovine milk samples in small-scale urban and peri-urban farming in Tajikistan was reported to have been 10.3% in seropositive and seronegative cows. Two individual strains, one as B. abortus and one as B. melitensiswere recognized (14). This proportion was 7.1% in Samsun, and 95% in Erzurum, Turkey, with all the strains identified as B. abortus, respectively (44,46). It is noteworthy to state that since vaccination against brucellosis is infrequently applied in Mediterranean and Central Asian countries, this may clarify the high rate of the infection in some cases (14,47).
Conclusions
Holistically, given the evidence regarding the contamination of milk with zoonotic species of Brucella in Kurdistan province of Iran, continuous vaccination of both industrialized and traditional rearing ruminants, test and slaughter policy, accurate evaluation of the contamination status of milk, and encouraging the consumption of pasteurized milk and dairy products may warrant the protection against brucellosis or Malta fever in the region. Moreover, it is highly recommended to use Rev1 as well as RB51 for vaccinating cows.
Conflict of interests
The authors declare that there is no conflict of interests.
Acknowledgements
The authors wish to express special thanks to the officials and staffs of Islamic Azad University, Sanandaj Branch, Iran.
Ethical Approval
The samples were taken from the animals with official permission and under the supervision of the Institutional Animal Ethics and Research Committee of Islamic Azad University, Sanandaj Branch (Certificate No. 98/101/96500).
Authors’ contribution
This study was conceived by EA and the Sampling was performed by MHA. EA and MHA carried out the laboratory works and EA wrote this article. All the authors have read and approved the manuscript.
Funding
This work was not upheld by any foundation.
References
- Cardoso PG, Macedo GC, Azevedo V, Oliveira SC. Brucella spp noncanonical LPS: structure, biosynthesis, and interaction with host immune system. Microb Cell Fact 2006; 5:13. doi: 10.1186/1475-2859-5-13 [Crossref] [ Google Scholar]
- WHO. The control of neglected zoonotic diseases: A route to poverty alleviation. Report of a joint WHO/DFID-AHP meeting. Geneva. World Health Organization; 2005. http://www.who.int/zoonoses/Report_Sept06.pdf.
- Scholz HC, Revilla-Fernández S, Dahouk SA, Hammerl JA, Zygmunt MS, Cloeckaert A. Brucella vulpis sp nov, isolated from mandibular lymph nodes of red foxes (Vulpes vulpes). Int J Syst Evol Microbiol 2016; 66(5):2090-8. doi: 10.1099/ijsem.0.000998 [Crossref] [ Google Scholar]
- Godfroid J, Scholz HC, Barbier T, Nicolas C, Wattiau P, Fretin D. Brucellosis at the animal/ecosystem/human interface at the beginning of the 21st century. Prev Vet Med 2011; 102(2):118-31. doi: 10.1016/j.prevetmed.2011.04.007 [Crossref] [ Google Scholar]
- Corbel MJ. Brucellosis in humans and animals, World Health Organization in collaboration with the Food and Agriculture Organization of the United Nations and World Organization for Animal Health. 2006. http://www.who.int/csr/resources/publications/Brucellosis.pdf.
- Franco MP, Mulder M, Gilman RH, Smits HL. Human brucellosis. Lancet Infect Dis 2007; 7(12):775-86. doi: 10.1016/s1473-3099(07)70286-4 [Crossref] [ Google Scholar]
- Pappas G. The changing Brucella ecology: novel reservoirs, new threats. Int J Antimicrob Agents 2010; 36 Suppl 1:S8-11. doi: 10.1016/j.ijantimicag.2010.06.013 [Crossref] [ Google Scholar]
- Shafei B, Ahmadi M, Dastmalchi Saei H. Diagnosis of Brucella abortus and Brucella melitensis in the milk of cattle and sheep in Kordestan province by polymerase chain reaction. J Vet Microbiol 2012; 8(2):127-35. [ Google Scholar]
- Shams N, Jaidari A, Etemadfar L. Molecular detection of Brucella abortus and Brucella melitensis in raw and unpasteurized bulk cow milk tanks of traditional domestic dairy sale centres in Khorramabad. Iran J Med Microbiol 2017; 11(4):13-20. [ Google Scholar]
- Al Dahouk S, Tomaso H, Nöckler K, Neubauer H, Frangoulidis D. Laboratory-based diagnosis of brucellosis--a review of the literature Part II: serological tests for brucellosis. Clin Lab 2003; 49(11-12):577-89. [ Google Scholar]
- Mailles A, Rautureau S, Le Horgne JM, Poignet-Leroux B, d’Arnoux C, Dennetière G. Re-emergence of brucellosis in cattle in France and risk for human health. Euro Surveill 2012; 17(30):20227. [ Google Scholar]
- Muñoz PM, Marín CM, Monreal D, González D, Garin-Bastuji B, Díaz R. Efficacy of several serological tests and antigens for diagnosis of bovine brucellosis in the presence of false-positive serological results due to Yersinia enterocolitica O:9. Clin Diagn Lab Immunol 2005; 12(1):141-51. doi: 10.1128/cdli.12.1.141-151.2005 [Crossref] [ Google Scholar]
- Godfroid J, Al Dahouk S, Pappas G, Roth F, Matope G, Muma J. A “One Health” surveillance and control of brucellosis in developing countries: moving away from improvisation. Comp Immunol Microbiol Infect Dis 2013; 36(3):241-8. doi: 10.1016/j.cimid.2012.09.001 [Crossref] [ Google Scholar]
- Lindahl-Rajala E, Hoffman T, Fretin D, Godfroid J, Sattorov N, Boqvist S. Detection and characterization of Brucella spp in bovine milk in small-scale urban and peri-urban farming in Tajikistan. PLoS Negl Trop Dis 2017; 11(3):e0005367. doi: 10.1371/journal.pntd.0005367 [Crossref] [ Google Scholar]
- Zambriski JA, Maves RC, Nydam DV, Ayvar V, Cepeda D, Castillo R. Effect of storage temperature and sample volume on Brucella melitensis isolation from goat milk. Int J Trop Dis Health 2012; 2(3):207-13. doi: 10.9734/ijtdh/2012/1738 [Crossref] [ Google Scholar]
- Quinn PJ, Carter ME, Markey B, Carter GR. Clinical Veterinary Microbiology. Ireland: Mosby Publisher; 2004.
- Baily GG, Krahn JB, Drasar BS, Stoker NG. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg 1992; 95(4):271-5. [ Google Scholar]
- Whatmore AM, Perrett LL, MacMillan AP. Characterisation of the genetic diversity of Brucella by multilocus sequencing. BMC Microbiol 2007; 7:34. doi: 10.1186/1471-2180-7-34 [Crossref] [ Google Scholar]
- Bricker BJ, Halling SM. Differentiation of Brucella abortus bv 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv 1 by PCR. J Clin Microbiol 1994; 32(11):2660-6. doi: 10.1128/jcm.32.11.2660-2666.1994 [Crossref] [ Google Scholar]
- Ahmadi E. Potential public health risk due to consumption of contaminated bovine milk with aflatoxin M1 and Coxiella burnetii in the West of Iran. Int J Dairy Technol 2020; 73(3):479-85. doi: 10.1111/1471-0307.12687 [Crossref] [ Google Scholar]
- Ahmadi E, Mardani K, Amiri A. Molecular detection and antimicrobial resistance patterns of Shiga toxigenic Escherichia coli isolated from bovine subclinical mastitis milk samples in Kurdistan, Iran. Arch Razi Inst 2020; 75(2):169-77. doi: 10.22092/ari.2019.124238.1278 [Crossref] [ Google Scholar]
- Khazaie F, Ahmadi E. Bovine subclinical mastitis-associated methicillin-resistant Staphylococcus aureus, selective genotyping and antimicrobial susceptibility profile of the isolates in Kurdistan province of Iran. Iran J Microbiol 2021; 13(1):65-73. doi: 10.18502/ijm.v13i1.5494 [Crossref] [ Google Scholar]
- Ocholi RA, Kwaga JK, Ajogi I, Bale JO. Phenotypic characterization of Brucella strains isolated from livestock in Nigeria. Vet Microbiol 2004; 103(1-2):47-53. doi: 10.1016/j.vetmic.2004.06.012 [Crossref] [ Google Scholar]
- Esmaeili S, Pourhossein B, Gouya MM, Bagheri Amiri F, Mostafavi E. Seroepidemiological survey of Q fever and brucellosis in Kurdistan province, western Iran. Vector Borne Zoonotic Dis 2014; 14(1):41-5. doi: 10.1089/vbz.2013.1379 [Crossref] [ Google Scholar]
- Hosseini S, Tanomand A, Rajabzadeh R, Ahmadpour M. Epidemiological aspects of brucellosis in Bane county during 2011-2012. J North Khorasan Uni Med Sci 2016; 7(3):485-94. doi: 10.29252/jnkums.7.3.485.[Persian] [Crossref] [ Google Scholar]
- Norouzinezhad F, Erfani H, Norouzinejad A, Kaveh F, Ghaffari F. Epidemiological characteristics and trend of the incidence of human brucellosis in Kurdistan province from 2009 to 2016. Iran J Epidemiol 2020; 15(4):323-33. [ Google Scholar]
- Gardner IA, Stryhn H, Lind P, Collins MT. Conditional dependence between tests affects the diagnosis and surveillance of animal diseases. Prev Vet Med 2000; 45(1-2):107-22. doi: 10.1016/s0167-5877(00)00119-7 [Crossref] [ Google Scholar]
- Godfroid J, Nielsen K, Saegerman C. Diagnosis of brucellosis in livestock and wildlife. Croat Med J 2010; 51(4):296-305. doi: 10.3325/cmj.2010.51.296 [Crossref] [ Google Scholar]
- Godfroid J, Saegerman C, Wellemans V, Walravens K, Letesson JJ, Tibor A. How to substantiate eradication of bovine brucellosis when aspecific serological reactions occur in the course of brucellosis testing. Vet Microbiol 2002; 90(1-4):461-77. doi: 10.1016/s0378-1135(02)00230-4 [Crossref] [ Google Scholar]
- Gwida M, El-Ashker M, Melzer F, El-Diasty M, El-Beskawy M, Neubauer H. Use of serology and real time PCR to control an outbreak of bovine brucellosis at a dairy cattle farm in the Nile Delta region, Egypt. Ir Vet J 2015; 69:3. doi: 10.1186/s13620-016-0062-9 [Crossref] [ Google Scholar]
- Office International des Epizooties (OIE). Bovine Brucellosis; caprine and ovine brucellosis and porcine brucellosis. In: World assembly of delegates of the OIE chapter 2.4.3. Paris: OIE Terrestrial Manual; 2009. p. 1-35.
- Rijpens NP, Jannes G, Van Asbroeck M, Rossau R, Herman LM. Direct detection of Brucella spp in raw milk by PCR and reverse hybridization with 16S-23S rRNA spacer probes. Appl Environ Microbiol 1996; 62(5):1683-8. doi: 10.1128/aem.62.5.1683-1688.1996 [Crossref] [ Google Scholar]
- Dadar M, Alamian S, Behrozikhah AM, Yazdani F, Kalantari A, Etemadi A. Molecular identification of Brucella species and biovars associated with animal and human infection in Iran. Vet Res Forum 2019; 10(4):315-21. doi: 10.30466/vrf.2018.89680.2171 [Crossref] [ Google Scholar]
- Ashrafganjooyi SH, Saedadeli N, Alamian S, Khalili M, Shirazi Z. Isolation and biotyping of Brucella spp from sheep and goats raw milk in southeastern Iran. Trop Biomed 2017; 34(3):507-11. [ Google Scholar]
- Tekle M, Legesse M, Edao BM, Ameni G, Mamo G. Isolation and identification of Brucella melitensis using bacteriological and molecular tools from aborted goats in the Afar region of north-eastern Ethiopia. BMC Microbiol 2019; 19(1):108. doi: 10.1186/s12866-019-1474-y [Crossref] [ Google Scholar]
- Al-Mariri A. Isolation of Brucella melitensis strains from Syrian bovine milk samples. Bulg J Vet Med 2015; 18(1):40-8. doi: 10.15547/bjvm.842 [Crossref] [ Google Scholar]
- Marin CM, Jiménez de Bagüés MP, Barberán M, Blasco JM. Comparison of two selective media for the isolation of Brucella melitensis from naturally infected sheep and goats. Vet Rec 1996; 138(17):409-11. doi: 10.1136/vr.138.17.409 [Crossref] [ Google Scholar]
- Shirazi Z, Khalili M, Sadeghi B, Sharifi H, Ashrafganjooyi S. Detection of Brucella spp in the sheep and goats milks from southeastern Iran using culture and PCR. J Med Microbiol Infect Dis 2017; 5(3):40-2. doi: 10.29252/JoMMID.5.3.4.40 [Crossref] [ Google Scholar]
- Doosti A, Ghasemi Dehkordi P. Application of real-time PCR for identification and differentiation of Brucella abortus and Brucella melitensis in cattle. Bulg J Vet Med 2011; 14(2):109-15. [ Google Scholar]
- Zowghi E, Ebadi A. Abortion due to Brucella abortus in sheep in Iran. Rev Sci Tech 1988; 7(2):379-82. doi: 10.20506/rst.7.2.356 [Crossref] [ Google Scholar]
- Zowghi E, Ebadi A, Yarahmadi M. Isolation and identification of Brucella organisms in Iran. Iran J Clin Infect Dis 2008; 3(4):185-8. [ Google Scholar]
- Romero C, Pardo M, Grillo MJ, Diaz R, Blasco JM, Lopez-Goñi I. Evaluation of PCR and indirect enzyme-linked immunosorbent assay on milk samples for diagnosis of brucellosis in dairy cattle. J Clin Microbiol 1995; 33(12):3198-200. doi: 10.1128/jcm.33.12.3198-3200.1995 [Crossref] [ Google Scholar]
- Moradi G, Esmaiel Nasab N, Ghaderi E, Sofi Majidpour M, Salimzadeh H. Brucellosis in Kurdistan province from 1997 to 2003. Annals of Alquds Medicine 2006; 2(1):32-7. [ Google Scholar]
- Shakerian A. Study of contamination rate in craw milk and its traditional products with Brucella abortus, and Brucella mellitensis in Isfahan and Chaharmahal and Bakhtiari provinces, 2012. J Shahrekord Uun Med Sci 2015; 17(1):16-23. [ Google Scholar]
- Esmaeili H, Ekhtiyarzadeh H, Ebrahimzadeh H, Partovi R, Marhamati Khamemeh B, Hamedi M. Evaluation of the national sheep and goat brucellosis control program in Iran. J Arak Uni Med Sci 2012; 14(7):9-20. [ Google Scholar]
- Terzi̇ G, Büyüktanir Ö, Genc O, Gücükoğlu A, Yurdusev N. Detection of Brucella antibody and DNA in cow milk by ELISA and PCR methods. Kafkas Univ Vet Fak Derg 2010; 16(Suppl A):S47-52. doi: 10.9775/kvfd.2009.1284 [Crossref] [ Google Scholar]
- Arasoğlu T, Güllüce M, Özkan H, Adigüzel A, Şahin F. PCR detection of Brucella abortus in cow milk samples collected from Erzurum, Turkey. Turk J Med Sci 2013; 43(4):501-8. doi: 10.3906/sag-1205-121 [Crossref] [ Google Scholar]