Avicenna Journal of Clinical Microbiology and Infection. 8(1):27-33.
doi: 10.34172/ajcmi.2021.06
Original Article
Evaluation of the Antibacterial and Investigation of the Molecular Docking of New Derivatives of 1, 3, 4-Oxadiazole as Inhibitors of Quorum Sensing System in the Human Pathogen Pseudomonas Aeruginosa
Sepideh Ghameshlouei 1  , Nakisa Zarrabi Ahrabi 1
, Nakisa Zarrabi Ahrabi 1  , Ali Souldozi 2
, Ali Souldozi 2  , Yasin SarveAhrabi 1, *
, Yasin SarveAhrabi 1, * 
Author information:
1Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran
2Department of chemistry, Urmia Branch, Islamic Azad University, Urmia, Iran
*
Corresponding author: Yasin SarveAhrabi, PhD in Microbiology, Department of Biology, Central Tehran Branch, Islamic Azad University, Tehran, Iran, Tel: +989141483263, Email:
yasin.ahrabi2016@gmail.com
Abstract
Background: Oxadiazoles are a group of anti-inflammatory compounds that have a wide range of activity due to their higher efficacy. Pseudomonas aeruginosa is an opportunistic pathogen and a major pathogen of nosocomial infections. This study aimed to evaluate the antibacterial and investigation of the molecular docking of new derivatives of 1, 3, 4-oxadiazole against P. aeruginosa, in vitro & in silico.
Materials and Methods: Four new derivatives were synthesized and added to our previous synthetic derivatives of 1, 3, 4-oxadiazole. The antibacterial activity of all derivatives was measured based on three standard species of P. aeruginosa using inhibition zone (IZ) and minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) methods. Then, employing the computational design of the drug by the molecular docking method, the inhibitory effect of synthetic compounds on the LasR regulatory protein of P. aeruginosa quorum sensing system was investigated, which plays an important role in regulating the expression of pathogenic genes in bacteria.
Results: The chemical structures of new compounds were characterized by IR spectra and 1H-NMR. A variety of inhibitory effects were observed by the synthesized compounds – compound 4d and 4g, in particular. Also, the inhibitory effect of these two compounds on the LasR regulatory protein under the control of the quorum sensing system in P. aeruginosa was demonstrated by molecular docking.
Conclusions: The results of this study showed that the two compounds containing the functional group of naphthalene and fluorophenyl have a significant effect on the inhibition of P. aeruginosa, as well as on the LasR protein of this bacterium.
Keywords: Oxadiazoles, Antibacterial, Molecular docking, Fluorophenyl, Naphthalene
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Pseudomonas aeruginosa is a gram-negative bacillus without fermentation power, opportunistic with aggressive power that is resistant to antimicrobial agents (1). Increased antibiotic resistance of this bacterium is a major health problem (2). It is the natural flora of the skin and intestines that causes respiratory infections, urinary tract infections, gastrointestinal infections, keratitis, otitis and bacteremia in patients with weakened immune systems (such as cancer, burns, AIDS, cystic fibrosis). These infections often cause death in patients (3). Nowadays, one of the most important issues in public health is the emergence of antibiotic-resistant strains and antimicrobial compounds in various communities, especially in hospitals (4). Addressing this issue requires the design and synthesis of newer and more effective drug compounds, and it is becoming more and more important (5). Due to the clinical importance and increasing drug resistance of P. aeruginosa infection caused by a variety of virulence factors, high antibiotic resistance and biofilm formation, the present study attempted to investigate the inhibitory effect of new 1, 3, 4-oxadiazole compounds on the quorum sensing system in P. aeruginosa. Bacteria have a system called quorum sensing, which is a cell-to-cell communication mechanism in bacteria and is involved in the development of infectious diseases by pathogenic bacteria through synthesizing and expressing pathogenicity indicators. The quorum sensing system is of the LuxR type and is controlled by self-propagating signal molecules called inducers. Signal molecules are produced naturally as bacteria grow. Over time, as the number of bacteria increases, the amount of these signal molecules or self-inducers also increases in the environment and as soon as they reach the threshold by activating the target genes dependent on the quorum sensing system, they induce and express certain phenotypic effects (6). Oxadiazoles is one of the structures considered by researchers for synthesizing new therapeutic compounds (7). There are four known isomers of this 5-membered heterocycle, including 1, 2, 4-oxadiazole 1, 2, 3-oxadiazole 1, 2, 5-oxadiazole, and 1, 3, 4-oxadiazole (8). 1, 3, 4-oxadiazoles are of special importance due to their prominent biological properties such as antibacterial (7), antifungal (9), anti-tuberculosis (10) and anti-cancer (11) properties. The introduction of 1, 3, 4-oxadiazole ring to the inhibitors can change their polarity and flexibility, as well as their metabolic stability (12). As the acceptor of hydrogen bonds formation, meanwhile, the 1, 3, 4-oxadiazole scaffold has the potential to be an isosteric substituent for amide or ester groups, so as to interfere protein or lipid biosynthesis in pathogens (13). Given the fact that bioinformatics and information technology are advancing today at an unexpected pace and their importance cannot be denied in the fields of medicine, biotechnology, and pharmacology, now along with traditional methods of drug synthesis, computer-synthesized structures are also approved in terms of antibacterial power through computer calculations with the least cost, which, if appropriate, lead to the therapeutic use (14). In the present study, the synthesis of new 1, 3, 4-oxadiazole compounds along with our previous derivatives, and the inhibitory effect of these compounds on three different strains of P. aeruginosa and LasR protein (chain H) as the most important regulatory protein in the quorum sensing system of this bacterium were investigated experimentally and computationally.
Methods
Chemicals
Previous synthetic structures of 1, 3, 4-oxadiazoles were re-synthesized (15).
All beginning and lawful materials were prepared from Merck Company (Germany) and used moving forward without any more filtration. Infrared spectrum was measured by a Shimadzu IR-460 spectrometer. Nuclear magnetic resonance spectrum was obtained by a Bruker DRX-300 AVANCE spectrometer (1H NMR at 300 Hz) in CDCl3. Chromatography columns were prepared using silica gel powder (Merck, Germany).
One-step Process for the Synthesis of 1, 3, 4-Oxadiazole Derivatives (4j-o)
First 0.01 mL of 2-pyridine-carbaldehyde was dissolved with 0.07 mL of di-ethyl-amine in acetonitrile, which was used as a solvent. After stirring vigorously for 5 minutes, 0.03 g of N-iso-cyano-imino-triphenyl-phosphorane was slowly added. TLC showed that after 2 hours the reaction was complete and the product was formed and was separated by a plate with a ratio of 10:6 (petroleum ether to ethyl acetate). After evaporation of the solvent, the product was obtained (15).
Antimicrobial Assay
Microorganism
Culture mediums (Mueller-Hinton agar, Mueller-Hinton Broth) were obtained from Merck Company (Germany). P. aeruginosa strains were prepared from the Iranian Industrial Microorganisms Collection Center (Lyophilized). Microbiological tests were performed using a Memmert- INC153T2T3 incubator.
Strains: P. aeruginosa (1): PTCC No: 1707/ATCC: 15442/ CIP: 103467/ NCIB: 10421
P. aeruginosa (2): PTCC No: 1599/ NCIMB: 10817/ ATCC: 25619/ FDA strain PCI: 852
P. aeruginosa (3): PTCC No: 1430/ ATCC: 27853/ DSM: 1117/ CCUG: 17619/ CECT: 108
inhibition zone and Broth microdilution (minimum inhibitory concentration and minimum bactericidal concentration) were applied to evaluate antibacterial susceptibility tests (14). The results were reported as the ±mean of three independent experiments.
Inhibition Zone
After preparing the suspension of each strain of bacteria in the tube with distilled water and matching the turbidity of the suspensions with the turbidity equivalent to 0.5 McFarland standards (1.5 × 108 CFU/mL), some of each suspension were removed with sterile cotton and were pure culture culture in Müller Hinton agar medium. Then, wells were made in the plate by Pasteur pipette. Then, 10 μL of solutions prepared from 1, 3, 4-oxadiazole derivatives (at concentrations of 0.5 mg/mL) were injected into the wells. Finally, the plates were closed and they were incubated for 24 hours at 37°C. After the mentioned time, the plates were examined for the presence of growth inhibition zone. It should be noted that ciprofloxacin (at concentrations of 0.5 mg/mL) was considered as a control sample (15).
Minimum Inhibitory Concentration Experiment
Minimum inhibitory concentration of the compounds was determined using dilution agar method. Concentrations of 1000, 500, 250, 125, 62.50, 31.25, 15.62 µg/mL were prepared in sterile molten Muller Hinton Broth from the stock solution. The microplates were inoculated with 1.5×106 cfu/mL of bacterial suspension at 37°C for 24 hours. The MIC was defined as the lowest concentration of the compounds that prevented visible growth of microorganisms after 24h incubation. The results were compared with ciprofloxacin (at concentrations of 0.5 mg/mL) as standard antibacterial agents. Stock solutions of 1, 3, 4-oxadiazole derivatives were prepared at concentration of 0.5 mg/mL in dimethyl sulfoxide (0.001 g of each derivative in 2 cc of DMSO). The lowest concentration that inhibited growth of bacteria was defined as the MIC (16). All experiments were performed in duplicate and results were reported as mean ± standard deviation.
MBC Experiment
MBC refers to the lowest concentration of antibiotic that can reduce the bacterial population by 9.99% after 24 hours; in other words, it can reduce the initial population a thousand times more. After determining the MIC, the tubes are cultured on a sterile Müller Hinton agar culture medium with a swap, which is placed in a 37°C incubator for 24 hours (17).
Molecular Docking
Protein Preparation
The crystallographic structure of the LasR protein complex of P. aeruginosa with a resolution of 1.83 angstroms was downloaded from the protein database (PDB ID: 2UV0). By removing all ligand molecules, waters, excess molecules, as well as the chains of protein that were not required for docking and then by adding polar hydrogen and charge to it, the desired protein was prepared for the docking process. Chain (H) was used for this docking (Figure 1).
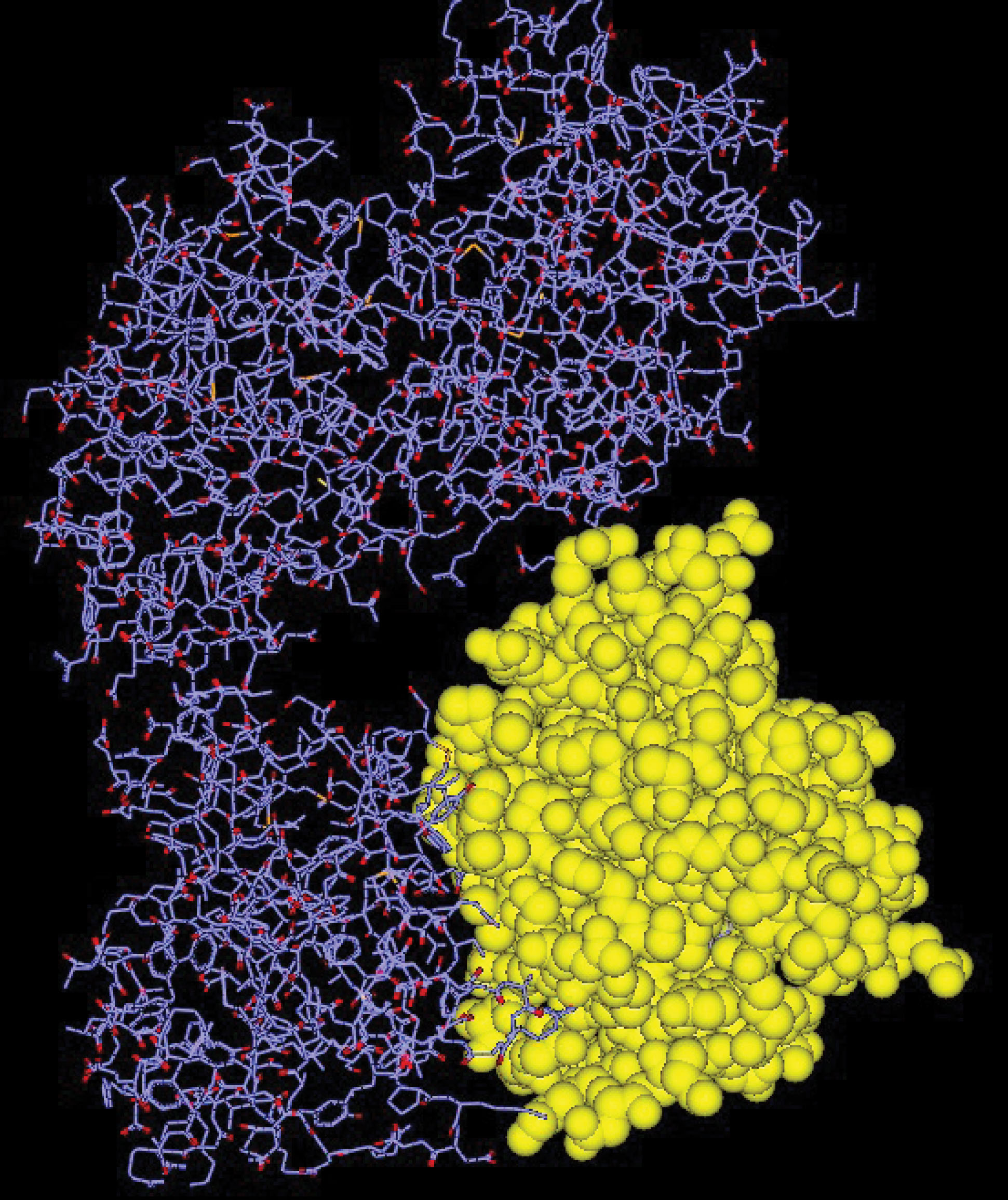
Figure 1.
Structure of the P. aeruginosa LasR Ligand-Binding Domain (Chain H= Yellow).
.
Structure of the P. aeruginosa LasR Ligand-Binding Domain (Chain H= Yellow).
Ligand Preparation
The fourteen compounds of 1, 3, 4-oxadiazole, that are called ligands in this program, were plotted by professional ChemDraw software (19.1) and then the energy optimization of structures using molecular mechanical (MM +) and quasi-experimental methods ( AM1) was applied by Chem3D (19.1).
Docking
Using the AutoDockTools (1.5.6) program, all hydrogen atoms were added to the protein structure, and the partial loads of the ligand were calculated and added with Discovery Studio 4.5 Client program; Grid box being 40×40×40 dimensional and Dimensions were also considered as X-center = 26.668, Y-center = 32.892 and Z-center = 41.388. Then using AutoDock Vina (18), the best format with the least amount of connection energy was selected as the stable format and the best combination in terms of (-ΔG) was examined by Discovery Studio (4.5 Client) software (2D and 3D).
Results
Chemicals
The structures (4a-4o), infrared and NMR of all compounds (4j-4o) were obtained (Table 1). Our study is novel in chemistry since it investigated the synthesis of 4j-4o structures for the first time in the form of single-stage synthesis. The results of the study showed that the synthetic products could be obtained with a purity of over 95%. To continue the experiments, all products were used as liquid solutions at a concentration of 0.5 mg/mL. Chemical formula, exact mass and molecular weight of all compounds were calculated with chem3D (Table 2).
Table 1.
The Final Structure of Compounds and their Spectral Information
|
Compound
|
Name and Structure
|
| 4a* |
(5-(Ph)-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4b* |
(5-(3-(Br-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4c* |
(5-(3-(Cl-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4d* |
(5-((N)-2-il)-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4e* |
(5-(3-( Fl-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4f* |
(5-(4-( Fl-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4g* |
(5-(3,4-(Di- Fl-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4h* |
(5-(4-(H3CO-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4i* |
(5-(3-(H3CO-Ph))-1,3,4-Oxadiazole-2-il)(Pyridine-2-il)Methanol
|
| 4j |
N-ethyl-N-((5-(3-Chlorophenyl)-1, 3, 4-oxadiazol-2-yl)(pyridin-2-yl)methyl)piperidine |
|
|
IR (KBr) (νmax, cm-1): 3433, 2846, 1667, 1536, 1492, 1378, 1289, 1046, 922.
1HNMR (300.13 MHz): δH= 3.93 (3H, Cl, s),7.21(1H, CH), 6.98 (1H, OH), 7.20-7.23 (H, CH), 7.49 (1H= 3JHH), 7.64-7.68 (H, CH), 7.90-7.98 (H, CH), 7.84 (H= 3JHH, CH), 8 (2H, 3JHH, CH), 8.51 (1H, 3JHH, CH), 8.92-8.94 (H, CH) in CDCl3.
|
|
4k
|
N-ethyl-N-((5-(4-Chlorophenyl)-1, 3, 4-oxadiazol-2-yl)(pyridin-2-yl)methyl)piperidine |
|
|
IR (KBr) (νmax, cm-1): 3633, 3122, 2823, 2632, 1777, 1702, 1636, 1588, 1485, 1412, 1392, 1281, 1220, 958.
1HNMR (300.13 MHz): δH= 4.56 (3H, Cl, s),8.21(1H, CH), 7.98 (1H, OH), 8.20-8.23 (H, CH), 8.49 (1H= 3JHH), 8.64-8.68 (H, CH), 8.90-8.98 (H, CH), 8.84 (H= 3JHH, CH), 9 (2H, 3JHH, CH), 9.51 (1H, 3JHH, CH), 9.92-9.94 (H, CH) in CDCl3.
|
|
4l
|
N-ethyl-N-((5-(3-Fluorophenyl)-1, 3, 4-oxadiazol-2-yl)(pyridin-2-yl)methyl)piperidine |
|
|
IR (KBr) (νmax, cm-1): 3636, 3128, 2827, 2638, 1782, 1712, 1639, 1599, 1498, 1414, 1308, 1245, 1202, 974.
1HNMR (300.13 MHz): δH= 4.66 (3H, Fl, s),8.31(1H, CH), 7.99 (1H, OH), 8.30-8.33 (H, CH), 8.59 (1H= 3JHH), 8.74-8.78 (H, CH), 8.92-8.99 (H, CH), 8.78 (H= 3JHH, CH), 8.96 (2H, 3JHH, CH), 9.31 (1H, 3JHH, CH), 9.92-9.94 (H, CH) in CDCl3.
|
|
4m
|
N-ethyl-N-((5-(4-Fluorophenyl)-1, 3, 4-oxadiazol-2-yl)(pyridin-2-yl)methyl)piperidine |
|
|
IR (KBr) (νmax, cm-1): 3940, 3652, 2953, 2740, 1650, 1550, 1356, 1348, 1320, 1211, 1201, 980.
1HNMR (300.13 MHz): δH= 4.68 (3H, Fl, s),8.32(1H, CH), 7.90 (1H, OH), 8.31-8.34 (H, CH), 8.60 (1H= 3JHH), 8.874-8.78 (H, CH), 8.92-8.99 (H, CH), 8.78 (H= 3JHH, CH), 8.96 (2H, 3JHH, CH), 9.31 (1H, 3JHH, CH), 9.92-9.94 (H, CH) in CDCl3.
|
|
4n
|
N-ethyl-N-((5-(3-Bromophenyl)-1, 3, 4-oxadiazol-2-yl)(pyridin-2-yl)methyl)piperidine |
|
|
IR (KBr) (νmax, cm-1): 3531, 3223, 2828 2740, 1665, 1582, 1536, 1510, 1452, 1423, 1368, 1279, 1215, 1044.
1HNMR (300.13 MHz, CDCl3): δH= 3.93 (3H, Br, s), 6.14 (s, 1H, CH), 6.53 (s, 1H, OH), 7.15-7.19 (m, 1H, CH), 7.48 (t, 1H, 3JHH= 8.1 Hz), 7.63-7.66 (m, 1H, CH), 7.73-7.75 (m, 1H, CH), 7.83 ( d, 1H, 3JHH=8.1 Hz, CH), 7.99 (t, 2H, 3JHH =7.8 Hz, CH), 8.477 (d, 1H, 3JHH = 8.1 Hz, CH), 8.916-8.930 (m, 1H, CH).
|
|
4o
|
N-ethyl-N-((5-(4-Bromophenyl)-1, 3, 4-oxadiazol-2-yl)(pyridin-2-yl)methyl)piperidine |
|
|
IR (KBr) (νmax, cm-1): 3531, 3223, 2828 2740, 1665, 1582, 1536, 1510, 1452, 1423, 1368, 1279, 1215, 1044.
1HNMR (300.13 MHz, CDCl3): δH= 3.93 (3H, Br, s), 6.14 (s, 1H, CH), 6.53 (s, 1H, OH), 7.15-7.19 (m, 1H, CH), 7.48 (t, 1H, 3JHH= 8.1 Hz), 7.63-7.66 (m, 1H, CH), 7.73-7.75 (m, 1H, CH), 7.83 ( d, 1H, 3JHH=8.1 Hz, CH), 7.99 (t, 2H, 3JHH =7.8 Hz, CH), 8.477 (d, 1H, 3JHH = 8.1 Hz, CH), 8.916-8.930 (m, 1H, CH).
|
*Source: SarveAhrabi et al (15).
Table 2.
Chem3D and AutoDock Vina Results and Antibacterial Properties of 1, 3, 4-Oxadiazole Compounds (3a-o)
|
Compound
|
Chem3D
|
ADV
|
Pseudomonas aeruginosa
|
|
PTCC: 1707
|
PTCC: 1599
|
PTCC: 1430
|
|
CF
|
EM
|
MW
|
Affinity (kcal/mol)
|
IZ
|
MIC
|
MBC
|
IZ
|
MIC
|
MBC
|
IZ
|
MIC
|
MBC
|
|
4a*
|
C14H11N3O2
|
253/09 |
253/26 |
-6.6 |
11 ± 0.50 |
≤1000 |
1000 |
12 ± 0.50 |
≤1000 |
1000 |
11 ± 0.50 |
≤1000 |
1000 |
|
4b*
|
C14H10BrN3O2
|
331/00 |
332/16 |
-9.6 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
4c*
|
C14H10ClN3O2
|
287/05 |
287/70 |
-8.4 |
13 ± 0.50 |
≤1000 |
1000 |
13 ± 0.50 |
≤1000 |
1000 |
13 ± 0.50 |
≤1000 |
1000 |
|
4d*
|
C
18
H
13
N
3
O
2
|
303/10
|
303/32
|
-12.2**
|
34 ± 0.50
|
≤125
|
250
|
33 ± 0.50
|
≤125
|
250
|
36 ± 0.50
|
125
|
≤250
|
|
4e*
|
C14H10FN3O2
|
271/08 |
271/25 |
-9.9 |
16 ± 0.50 |
≤1000 |
1000 |
16 ± 0.50 |
≤1000 |
1000 |
17 ± 0.50 |
≤1000 |
1000 |
|
4f*
|
C14H10FN3O2
|
271/08 |
271/25 |
-10.7 |
16 ± 0.50 |
≤1000 |
1000 |
15 ± 0.50 |
≤1000 |
1000 |
16 ± 0.50 |
≤1000 |
1000 |
|
4g*
|
C
14
H
9
F
2
N
3
O
2
|
289/07
|
289/24
|
-11.1**
|
36 ± 0.50
|
125
|
≤250
|
36 ± 0.50
|
≤125
|
250
|
34 ± 1.50
|
≤125
|
250
|
|
4h*
|
C15H13N3O3
|
283/10 |
283/29 |
-10.8 |
15 ± 0.50 |
≤1000 |
1000 |
14 ± 0.50 |
≤1000 |
1000 |
14 ± 0.50 |
≤1000 |
1000 |
|
4i*
|
C15H13N3O3
|
283/10 |
283/29 |
-10.2 |
11 ± 0.50 |
≤1000 |
1000 |
12 ± 0.50 |
≤1000 |
1000 |
12 ± 0.50 |
≤1000 |
1000 |
|
4j
|
C19H19ClN4O
|
354/12 |
354/84 |
-10.0 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
4k
|
C19H19ClN4O
|
354/12 |
354/84 |
-10.2 |
15 ± 0.50 |
≤1000 |
1000 |
15 ± 0.50 |
≤1000 |
1000 |
17 ± 0.50 |
≤1000 |
1000 |
|
4l
|
C19H19FN4O
|
338/15 |
338/39 |
-6.9 |
15 ± 0.50 |
≤1000 |
1000 |
15 ± 0.50 |
≤1000 |
1000 |
17 ± 0.50 |
≤1000 |
1000 |
|
4m
|
C19H19FN4O
|
338/15 |
338/39 |
-10.3 |
14 ± 0.50 |
≤1000 |
1000 |
14 ± 0.50 |
≤1000 |
1000 |
12 ± 0.50 |
≤1000 |
1000 |
|
4n
|
C19H19BrN4O
|
398/07 |
399/29 |
-6.9 |
17 ± 0.50 |
≤1000 |
1000 |
17 ± 0.50 |
≤1000 |
1000 |
16 ± 0.50 |
≤1000 |
1000 |
|
4o
|
C19H19BrN4O
|
398/07 |
399/29 |
-6.7 |
16 ± 0.50 |
≤1000 |
1000 |
17 ± 0.50 |
≤1000 |
1000 |
12 ± 0.50 |
≤1000 |
1000 |
ADV: AutoDock Vina, CF: Chemical Formula, EM: Exact Mass, MW: Molecular Weight, IZ: mm, MIC and MBC: μg/mL.
* *Source: SarveAhrabi et al (15).
**Best affinity of compounds to use for docking.
In Vitro
According to Figure 2, which deals with Inhibition zone of compounds on the target bacterium, structures containing naphthalene (N) and difluorophenyl (Di- Fl-Ph) showed the best performance. These results are reported in Table 2. As Table 2 and Figure 2 show, it seems that the compound 4d, which has an aromatic hydrocarbon and is widely used for disinfection and insecticide (mostly dissolved in methanol), had a very favorable effect on the bacteria along with the oxadiazoles ring. Compound 4g, which has one of the aromatic compounds and by pairing the two aromatic compounds of fluoro, created a favorable property against the studied bacteria.
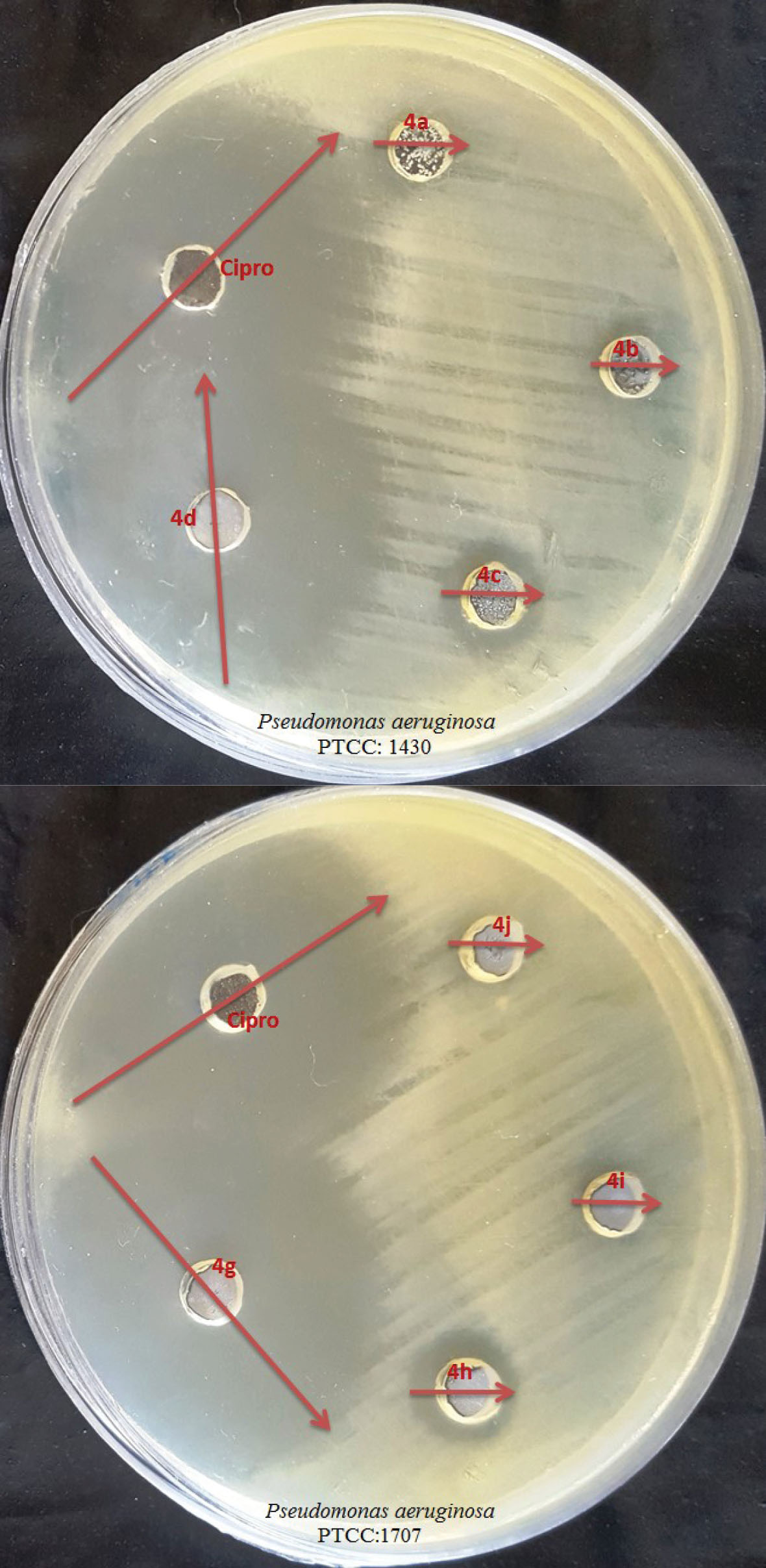
Figure 2.
Inhibition Zone of Compounds against P. aeruginosa at 0.5 Mg/Ml Concentration.
.
Inhibition Zone of Compounds against P. aeruginosa at 0.5 Mg/Ml Concentration.
Following Table 2, the effect of compound 4d was against P. aeruginosa with IZ= ± (35.00 ± 0.50) mm, MIC=125 mg/mL, and MBC=250 mg/mL. This result can be due to the presence of naphthalene group in the main compound (17). The effect of compound 4g was against P. aeruginosa with IZ= ± (35.00 ± 0.33) mm, MIC=125 mg/mL, and MBC = 250 mg/mL. This result may be due to the presence of Fluorophenyl group in the main compound with tow site (18).
In Silico
As shown in Table 2, all affinities of the compounds were calculated, and the best compounds with low ΔG (-ΔG) were selected to continue experiments and to investigate chemical interactions. According to Figure 3, the highest amount of hydrogen bonds was related to the 4d compound with the amino acid alanine by 127 hydrogen bonds, valine:76, leucine:110, tyrosine:64, asparagine:73, tryptophan:88 and tyrosine:115, among which the compound bonds were the case. The opinion can be considered by hydrogen bonding the site of alanine, leucine and tyrosine. With 4g compound, the highest amount of hydrogen bonds was related to alanine: 127, valine: 76, tryptophan: 64, asparagine: 73, leucine: 110, Tyrosine: 115. The opinion can be considered by hydrogen bonding the site of alanine, leucine and tyrosine.
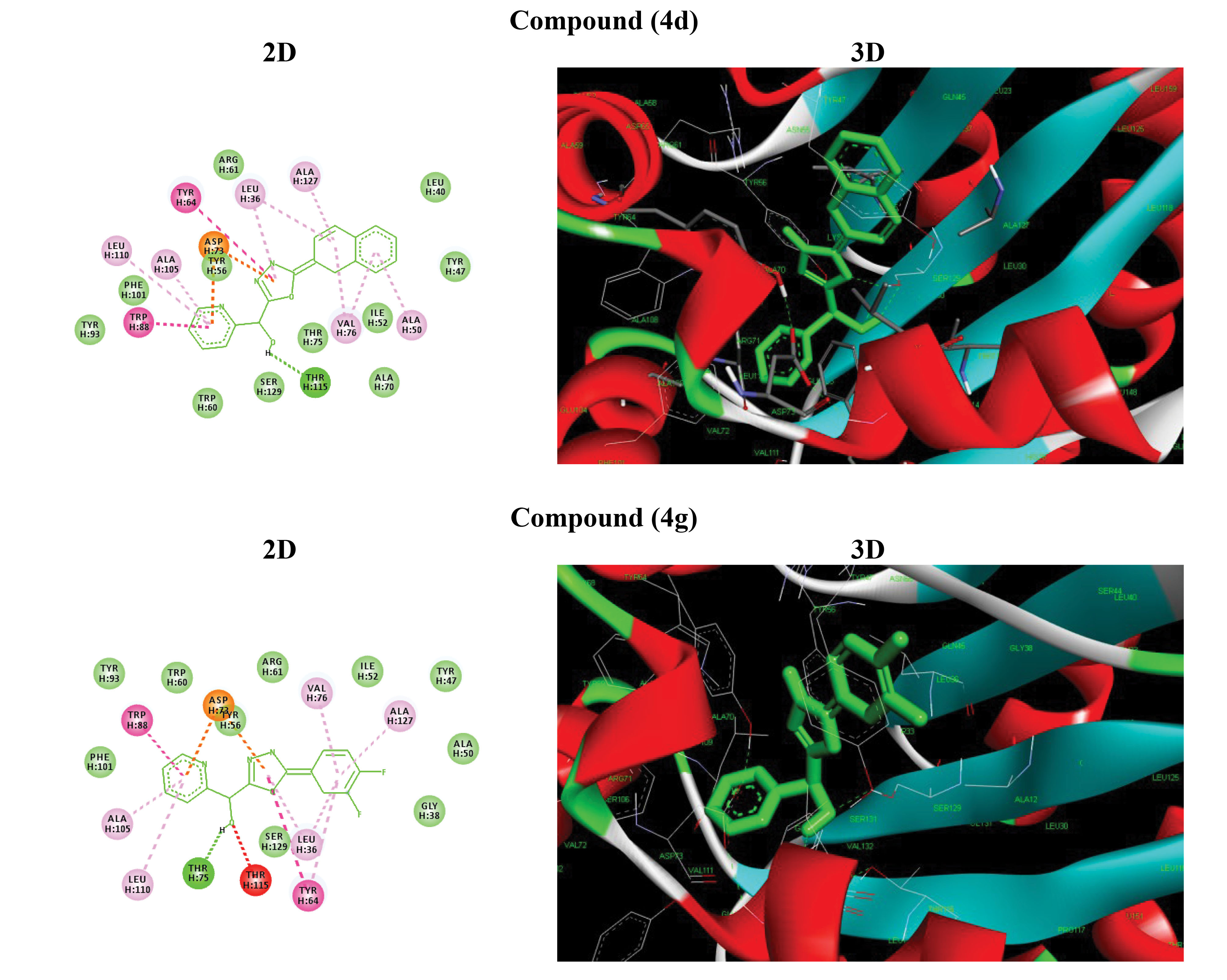
Figure 3.
Docked Conformation of Compound 4d and 4g in the Binding Site of LasR. Hydrogen Bonds Are Shown in 2D Figures.
.
Docked Conformation of Compound 4d and 4g in the Binding Site of LasR. Hydrogen Bonds Are Shown in 2D Figures.
Discussion
Generally, 1, 3, 4-oxadiazoles are multi-functionalized compounds with an assortment of organic properties and, therefore, their unused compound blend is vital. This study evaluated the antibacterial and in silico effects of 15 synthesized 1, 3, 4-oxadiazole derivatives (4j-4o new structures) against the Human Pathogen P. aeruginosa. These compounds, mainly Di-Fluorophenyl (4g) and Naphthalene (4d), showed relatively acceptable antibacterial effects against Gram-Negative P. aeruginosa. In a study conducted by Rayam et al, it was reported that the antibacterial activity of new synthesis compounds revealed 2‐(1‐(4‐isobutylphenyl)ethyl)‐5‐(((1‐phenyl‐1H‐1,2,3‐triazol‐4‐yl)methyl)thio)‐1,3,4‐oxadiazole (8b) and demonstrated more potent antibacterial activity against E. coli and P. aeruginosa (19). In another study, Chortani et al showed how synthesis of novel 1, 3, 4-oxadiazole linked benzopyrimidinones conjugates. Biological results of their study revealed that some of these compounds demonstrated excellent to moderate antimicrobial activities with minimum inhibitory concentration (MIC) values ranging from 10.8 to 140.7 μM. Notably, compounds 5g(1,4-dimethylbenzene) and 5h(4-chlorophenyl) showed the highest inhibitory activity against all bacterial strains with MIC values ranging from 111.3 to 10.8 μM against S. aureus, E. coli and P. aeruginosa (20). The results of these studies are consistent with the results of ours since the main role of the 1, 3, 4-oxadiazole ring has been mentioned in all given studies. In addition, in silico results indicated that Fluorophenyl ring and piridine-methanol ring in compound 4g play an important role in the formation of hydrogen bonds. Also, pirydine-methanol ring and oxadiazole ring with naphthalene in compound 4d are important factors in the formation of hydrogen bonds. Moreover, the simple workup, tall abdicates, and brief response times make the strategy a valuable expansion for planning advanced pharmaceutical synthetics. It is noteworthy that these derivatives (4j-4o) were synthesized for the first time and the relevant tests were performed to ensure the biological properties of the compounds. This research was done in order to provide new structures and confirm the existence of antibacterial activity and drug design (4a-4o) of these compounds.
Conclusions
It seems that 1, 3, 4-oxadiazole subordinates would be a supportive structure for conceivable improvement of unused drugs but this result must be affirmed by other broad clinical trials that would be a portion of our future plans.
Ethical Approval
The disclaimer implies that ethical principles have been considered in relation to the proposed work and no ethical issues have been found to be applied to this research proposal.
Conflict of Interests
The authors declare that they have no competing interests.
Acknowledgements
This research was the author’s project (Code: 162317565), done at the Islamic Azad University, Central Tehran Branch, Iran.
Authors’ Contributions
YS: Supervision, writing original draft, writing-reviewing, and editing; SG: writing original draft, data analysis; NZA: Data analysis; AS: Investigation, methodology.
Funding/Support
Islamic Azad University of Central Tehran Branch of Iran provided financial support.
References
- Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnology advances 2019 Jan 1; 37(1):177-92. [ Google Scholar]
- Pachori P, Gothalwal R, Gandhi P. Emergence of antibiotic resistance Pseudomonas aeruginosa in intensive care unit; a critical review. Genes & diseases 2019 Jun 1; 6(2):109-19. [ Google Scholar]
- Song D, Meng J, Cheng J, Fan Z, Chen P, Ruan H, Tu Z, Kang N, Li N, Xu Y, Wang X. Pseudomonas aeruginosa quorum-sensing metabolite induces host immune cell death through cell surface lipid domain dissolution. Nature microbiology 2019 Jan; 4(1):97-111. [ Google Scholar]
- Kageyama C, Sato M, Sakae H, Obayashi Y, Kawahara Y, Mima T, Matsushita O, Yokota K, Mizuno M, Okada H. Increase in antibiotic resistant Helicobacter pylori in a University Hospital in Japan. Infection and drug resistance 2019; 12:597. [ Google Scholar]
- Alharbi FA, Alarfaj AA. Green synthesis of silver nanoparticles from Neurada procumbens and its antibacterial activity against multi-drug resistant microbial pathogens. Journal of King Saud University-Science 2020 Mar 1; 32(2):1346-52. [ Google Scholar]
- Yu Z, Hu Z, Xu Q, Zhang M, Yuan N, Liu J, Meng Q, Yin J. The LuxI/LuxR-Type Quorum Sensing System Regulates Degradation of Polycyclic Aromatic Hydrocarbons via Two Mechanisms. International journal of molecular sciences 2020 Jan; 21(15):5548. [ Google Scholar]
- Zarrabi N, souldozi A, SarveAhrabi Y. In Vitro Evaluation of Antibacterial and Antifungal Properties of Some New 1, 3, 4-Oxadiazole Derivatives Containing Phenyl Group. IEM 2020; 6(3):177-192. [ Google Scholar]
- Boström J, Hogner A, Llinàs A, Wellner E, Plowright AT. Oxadiazoles in medicinal chemistry. Journal of medicinal chemistry 2012 Mar 8; 55(5):1817-30. [ Google Scholar]
- Capoci IR, Sakita KM, Faria DR, Rodrigues Vendramini FA, Arita GS, de Oliveira AG, Felipe MS, Maigret B, Bonfim-Mendonça PD, Kioshima ES, Svidzinski TI. Two new 1, 3, 4-oxadiazoles with effective antifungal activity against Candida albicans. Frontiers in microbiology 2019; 10:2130. [ Google Scholar]
- De SS, Khambete MP, Degani MS. Oxadiazole scaffolds in anti-tuberculosis drug discovery. Bioorganic & medicinal chemistry letters 2019 Aug 15; 29(16):1999-2007. [ Google Scholar]
- Sreenivasulu R, Tej MB, Jadav SS, Sujitha P, Kumar CG, Raju RR. Synthesis, anticancer evaluation and molecular docking studies of 2, 5-bis (indolyl)-1, 3, 4-oxadiazoles, Nortopsentin analogues. Journal of Molecular Structure 2020 May 15; 1208:127875. [ Google Scholar]
- Guimarães CR, Boger DL, Jorgensen WL. Elucidation of fatty acid amide hydrolase inhibition by potent α-ketoheterocycle derivatives from Monte Carlo simulations. Journal of the American Chemical Society 2005 Dec 14; 127(49):17377-84. [ Google Scholar]
- Rane RA, Gutte SD, Sahu NU. Synthesis and evaluation of novel 1, 3, 4-oxadiazole derivatives of marine bromopyrrole alkaloids as antimicrobial agent. Bioorganic & medicinal chemistry letters 2012 Oct 15; 22(20):6429-32. [ Google Scholar]
- Huang S, Ren Y, Peng X, Qian P, Meng L. Computer-aid drug design, synthesis, and anticoagulant activity evaluation of novel dabigatran derivatives as thrombin inhibitors. European Journal of Pharmaceutical Sciences 2019 Sep 1; 137:104965. [ Google Scholar]
- SarveAhrabi Y, Souldozi A, Zarrabi Ahrabi N. In Vitro Evaluation of Antimicrobial Properties of Some New 1, 3, 4-Oxadiazole Derivatives against Acinetobacter baumannii. Infection Epidemiology and Microbiology 2020 Feb 10; 6(1):37-49. [ Google Scholar]
- Magaldi S, Mata-Essayag S, De Capriles CH, Perez C, Colella MT, Olaizola C, Ontiveros Y. Well diffusion for antifungal susceptibility testing. International journal of infectious diseases 2004 Jan 1; 8(1):39-45. [ Google Scholar]
- Ikegbunam M, Ukamaka M, Emmanuel O. Evaluation of the antifungal activity of aqueous and alcoholic extracts of six spices. American Journal of Plant Sciences 2016 Jan 4; 7(1):118-25. [ Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry 2010 Jan 30; 31(2):455-61. [ Google Scholar]
- Rayam P, Polkam N, Kummari B, Banothu V, Gandamalla D, Yellu NR, Anireddy JS. Synthesis and Biological Evaluation of New Ibuprofen‐1, 3, 4‐oxadiazole‐1, 2, 3‐triazole Hybrids. Journal of Heterocyclic Chemistry 2019 Jan; 56(1):296-305. [ Google Scholar]
- Chortani S, Edziri H, Manachou M, Al-Ghamdi YO, Almalki SG, Alqurashi YE, Jannet HB, Romdhane A. Novel 1, 3, 4-oxadiazole linked benzopyrimidinones conjugates: Synthesis, DFT study and antimicrobial evaluation. Journal of Molecular Structure 2020 Oct 5; 1217:128357. [ Google Scholar]