Avicenna Journal of Clinical Microbiology and Infection. 8(2):57-65.
doi: 10.34172/ajcmi.2021.11
Original Article
Comparative Analysis of Prevalence and Antibiotic Resistance in Vancomycin-Resistant Enterococcus from Clinical Samples – Demographics and Phenotypes
Folasade Muibat Adeyemi 1, *  , Nana-Aishat Yusuf 1
, Nana-Aishat Yusuf 1  , Rashidat Ronke Adeboye 1
, Rashidat Ronke Adeboye 1  , Odunola Oluwaseun Oluwajide 1, Ajibade Kwashie Ako-Nai 2
, Odunola Oluwaseun Oluwajide 1, Ajibade Kwashie Ako-Nai 2
Author information:
1Department of Microbiology, Faculty of Basic and Applied Sciences, Osun State University, Osogbo, Nigeria
2Department of Microbiology, Faculty of Science, Obafemi Awolowo University, Ile-Ife, Osun State, Nigeria
*
Corresponding author: Folasade Muibat Adeyemi, Department of Microbiology, Faculty of Basic and Applied Sciences, Osun State University, Osogbo, Nigeria. Tel: +234 803 494 0747, Email:
folasade.adeyemi@uniosun.edu.ng
Abstract
Background: Of all enterococci species, the most renowned clinically as multidrug-resistant pathogens are Enterococcus faecium and Enterococcus faecalis. Vancomycin-resistant Enterococcus (VRE) species are the principal cause of opportunistic hospital-acquired infections, due to numerous resistance mechanisms.
Methods: In this study, the prevalence and antibiotic resistance profiles of VRE according to clinical sources from three selected hospitals in Southwest-Nigeria were investigated. Altogether, 431 samples (urine, rectal, and wound swabs - caesarian section (CS), automobile accidents, and other skin lesions and abrasions) were collected from three selected hospitals in Osun State, Nigeria. Established techniques were employed for the recovery of enterococci and screening for VRE while antibiotic susceptibility tests were carried out by disc diffusion technique.
Results: Altogether, 208 (48.3%) enterococci strains were recovered from which 85 (40.9%) were VRE. E. faecium predominated at 71.8% (61/85) and E. faecalis at 28.2% (24/85) as determined by phenotypic characterization. VRE isolates exhibited 100%, 97.6%, and 92.9% resistance to ampicillin, clindamycin, and quinupristin-dalfopristin (Q/D) respectively. The least resistance in-vitro was to tigecycline (27.1%). None of the antibiotics exhibited 100% activity against all the isolates. vanA resistant phenotype was prevalent at 65.9%. E. faecium from all study locations displayed higher levels of resistance than E. faecalis. Multiple antibiotic resistance (MAR) indices in all VRE isolates were ≥0.2, all being multidrug-resistant.
Conclusions: The high prevalence rate along with the high level of multidrug resistance observed in the present study is worrisome and poses a continuous threat in the therapy of illnesses triggered by VRE as vancomycin was perceived as a drug of choice to curb enterococcal infections.
Keywords: Enterococci, Vancomycin-resistance, Prevalence, Multidrug-resistance, Van phenotypes
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Enterococci constitute a significant portion of the natural gastrointestinal microbiome in both man and animal and can remain viable in varied forms of extreme environments. They are gram-positive cocci, facultative anaerobes, which commonly cause opportunistic nosocomial infections (1). Enterococcus faecium and Enterococcus faecalis reportedly being within the top three most significant nosocomial pathogens globally (2), however, are progressively more recognized and have emerged as clinically important multidrug-resistant infectious pathogens. They are implicated in many infections including bacteremia, endocarditis, urinary tract, intra-abdominal, pelvic, surgical site, and diabetic foot ulcer (3,4). This involvement could be due to their intrinsic resistance to different antibiotics, development of antibiotic resistance (2) with ability to adapt quickly in the health-care setting, and extreme genetic variations coupled with the incidence of a variety of virulence determinants (4), making enterococcal infections serious and life-threatening.
Enterococci species resistant to vancomycin (VRE) are purportedly a foremost source of opportunistic nosocomial infections (5). Multiple resistance mechanisms in VRE have led to limitations in available treatment options as increased vancomycin resistance in enterococci restricts the choice of vancomycin as a treatment for enterococcal infections (6). This is of public health importance as infections caused by VRE are challenging to manage in the clinical environment, and also the strains are capable of spreading between hospitals and districts (7). Increasing spread in the number of antimicrobial-resistant Enterococcus strains has been documented (8). Several new antimicrobials have been recently introduced and current information regarding combination therapies has shown promise, widening currently available options on therapy (5).
Resistance to vancomycin occurs mainly by acquiring the vanA and less frequently by the vanB gene, already described in detail in the specie E. faecium (9). Several risk factors exist for colonization by and consequent infection with vancomycin-resistant Enterococci. Predominant among these is prior exposure to antimicrobials (10), which possibly leads to bowel flora modification. Besides, patients with advancing age, immunocompromised states, and those with serious underlying illnesses – such as patients in institutions for long-term care, on prolonged antibiotics therapy, extended hospital stays, as well as proximity to other patients colonized with VRE – are at increased risk (11).
The part of enterococci in clinical infections has been insufficiently studied and reported in Nigeria. Previous reports have hinted that the development of resistance to glycopeptides among enterococci did not occur (12); but later, another study on the prevalence of VRE reported the detection of VRE from hospital samples, and hands of health care staff in Southwest Nigeria (13,14). Enterococcal isolates resistant to vancomycin are not habitually screened for in many clinical laboratories in Nigeria perhaps because of the perceived low incidence of resistance to vancomycin among enterococcal isolates, thereby obscuring the detection of vancomycin-resistance in enterococcal strains. Our study was therefore undertaken to evaluate the prevalence and antibiotic sensitivity pattern of vancomycin-resistant enterococci from clinical samples from three selected hospitals in Southwest Nigeria in order to influence treatment choices of infections triggered by VRE in the hospital environment.
Methods
Study Locations
This study was carried out at three designated hospitals - State Specialist Hospital, Osogbo (7.76958oN; 4.54999oE); Oke-Baale Primary Health Centre, Osogbo (7.76516oN; 4.578oE), and State Hospital, Iwo (7.66686oN; 4.19926oE), all located in Osun State, Southwest Nigeria. Osogbo, the capital city of Osun State with a projected population of 214 200 lies on coordinates 7.7827° N, 4.5418° E, while Iwo lies on coordinates 7.6292° N, 4.1872° E with 263 500 inhabitants.
Sample Collection and Processing
This was a cross-sectional study of urine, rectal swabs, and wound samples. The minimum sample size for the study (384 participants) was calculated with the formula n = z2pq /d2 where n is the least sample size mandatory, z is the centile of the standard normal distribution fixed at 1.96, p is the most probable prevalence rate of selected indicators (and in this study 50%), q is (1-p) and d is the degree of accuracy (at 0.05). Altogether, 431 participants were included in the study based on individual or parental consent for addition to the study.
The wound samples were obtained from wounds of caesarian sections (CS), trauma from automobile accidents, and other skin lesions and abrasions obtained from both out-patients and in-patients. Wound and rectal swabs were obtained with sterile swab-sticks dipped in sterile ringer solution, one swab per sample. The sterile swab-sticks were gently wiped on the exterior of the wound/rectum, then carefully rotated to sample the epithelial wall, removed, and inoculated aseptically into 10 mL of sterile Tryptone Soy Broth (TSB) (Oxoid, UK). The tubes were incubated at 37±2oC for 24 hours and then streaked onto Slanetz and Bartley agar. Urine samples collected mid-stream in sterile universal bottles were streaked out on Slanetz and Bartley agar and placed in the incubator at 37±2oC overnight. All samples were analyzed immediately upon arrival in the laboratory within 2 hours of collection. Pale pink or maroon colonies on Slanetz and Bartley agar were further identified by microscopic, morphologic, and biochemical characterization. Purecultures of the recovered isolates were stored in freshly prepared Tryptone Soy Broth supplemented with 15% glycerol and maintained at -20oC.
Identification on Bile Aesculin and Mannitol Salt Agar
Overnight growth from Slanetz and Bartley agar plates were inoculated onto Bile Aesculin Agar and Mannitol Salt Agar, then incubated at 37±2oC overnight. Growth on Bile Aesculin Agar indicated the capacity to grow with bile in the media, while a change in color of the agar to dark brown was indicative of aesculin hydrolysis, both being characteristic of enterococci species. E. faecalis grows and ferments mannitol on Mannitol Salt Agar (yellow colonies), while E. faecium lacks this trait (15).
Screening for Vancomycin Resistance
Vancomycin resistance was screened for with Brain Heart Infusion (BHI) Agar supplemented with 6 ug/mL vancomycin. Distinct colonies from an overnight culture on Nutrient agar were suspended in ringer solution, equivalent to 0.5 McFarland suspension, and spot-inoculated onto the screening agar. The plates were dried, placed in the incubator for 24 hours at 37±2°C in an inverted position. Growth of >1 colony indicated a vancomycin-resistant organism, while no growth denoted susceptibility to vancomycin.
Antibiotic Susceptibility Testing
Vancomycin-resistant enterococci were screened for sensitivity to other antibiotics using the Kirby-Bauer disc diffusion method. Antibiotics (Oxoid, UK) tested against the isolates were: ampicillin (10 µg), ciprofloxacin (5 µg), clindamycin (2 µg), gentamicin (10 µg), imipenem (10 µg), levofloxacin (1 µg), linezolid (30 µg), nitrofurantoin (100 µg), norfloxacin (10µg), quinupristin-dalfopristin (Q/D) (15 µg), teicoplanin (30 µg), tigecycline (15µg) and vancomycin (30 µg). The antibiotic discs were placed aseptically on Mueller Hinton Agar with an 8-place disc dispenser (Oxoid), and the petri dishes were incubated at 37±2°C overnight. The zones of inhibition were visually observed and measured to the nearest millimeter. The values were recorded and interpreted as susceptible, intermediate, or resistant using the EUCAST breakpoint tables version 9.0. Resistance to ≥1 antibacterial agent in ≥ 3 classes of antibiotics was used as an indicator of multidrug resistance (MDR)
Multiple antibiotic resistance (MAR) indices of recovered VRE species were estimated with the formula:
Results
Altogether, 431 patients participated in the study based on individual consent for inclusion (130 males, 30.2%; and 301 females, 69.8%). The age range of participants fell between 15 and 83 years (mean 33.3 years). A high percentage of participants (65.7%) were in the age bracket 21–40 years, eighty-two of who were male and 201 females. Details of the demographic information of the participants are shown in Table 1. Urine sample was the most prevalent sample collected – 62.4% (n=269/431), while wound swabs from CS constituted 4.2% (n=18/431) (Table 2).
Table 1.
Demographic Data of Participants
|
Criteria
|
Categories
|
Total (%)
|
Frequency
|
|
State Specialist Hospital, Osogbo (n=211)
|
Oke-Baale Primary Health Centre
(n=87)
|
State Hospital, Iwo (n=133)
|
|
Male (72)
|
Female (139)
|
Male (28)
|
Female (59)
|
Male (30)
|
Female (103)
|
| Age (y) |
≤20 |
44 (10.2) |
8 |
15 |
3 |
6 |
1 |
11 |
| 21-40 |
283 (65.7) |
45 |
92 |
17 |
40 |
20 |
69 |
| 41-60 |
87 (20.2) |
15 |
26 |
7 |
12 |
6 |
21 |
| 61-80 |
16 (3.7) |
4 |
5 |
1 |
1 |
3 |
2 |
| ≥81 |
1 (0.2) |
0 |
1 |
0 |
0 |
0 |
0 |
| Educational status |
Nil |
63 (14.6) |
14 |
13 |
5 |
8 |
7 |
16 |
| Primary |
57 (13.2) |
8 |
20 |
4 |
7 |
2 |
16 |
| Secondary |
104 (24.1) |
13 |
35 |
9 |
15 |
8 |
24 |
| Higher school |
135 (31.3) |
21 |
37 |
8 |
22 |
10 |
37 |
| Postgraduate |
72 (16.7) |
16 |
34 |
2 |
7 |
3 |
10 |
| Marital status |
Single |
138 (32.0) |
25 |
37 |
9 |
17 |
9 |
41 |
| Married |
293 (68.0) |
47 |
102 |
19 |
42 |
21 |
62 |
| Use of antibiotics |
On antibiotics |
206 (47.8) |
23 |
53 |
15 |
38 |
18 |
59 |
| Not on antibiotics |
225 (52.2) |
49 |
86 |
13 |
21 |
12 |
44 |
Table 2.
The Frequency of Occurrence of Recovered Isolates From Various Samples Based on Study Location and Gender of the Participants
|
Location
|
Sample Type
|
No. of Samples
|
No. of Enterococci
|
Patients With Enterococci
|
No. of VRE
|
Patients with VRE
|
|
Gender
|
Gender
|
Age Groups
|
|
Male
|
Female
|
Male
|
Female
|
≤ 20
|
21-40
|
41-60
|
61-80
|
≥ 81
|
State Specialist Hospital, Osogbo
(7.76958oN; 4.54999oE) |
Trauma |
49 |
25 |
7 |
18 |
14 |
7 |
7 |
3 |
9 |
2 |
0 |
0 |
| Skin lesions/abrasions |
24 |
15 |
9 |
6 |
10 |
1 |
9 |
2 |
6 |
1 |
1 |
0 |
| Caesarian section |
13 |
11 |
0 |
11 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Urine |
117 |
43 |
10 |
33 |
17 |
9 |
8 |
6 |
9 |
2 |
0 |
0 |
| Rectal swabs |
08 |
7 |
3 |
4 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Sub Total |
211 |
101 |
29 |
72 |
41 |
17 |
24 |
11 |
24 |
5 |
1 |
0 |
Oke-Baale Primary Health Centre
(7.76516oN; 4.578oE) |
Trauma |
03 |
3 |
2 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Skin lesions/abrasions |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
| Caesarian section |
02 |
1 |
0 |
1 |
1 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
| Urine |
68 |
31 |
25 |
6 |
10 |
2 |
8 |
0 |
6 |
4 |
0 |
0 |
| Rectal swabs |
14 |
5 |
2 |
3 |
1 |
1 |
0 |
0 |
1 |
0 |
0 |
0 |
| SUB TOTAL |
87 |
40 |
29 |
11 |
12 |
3 |
9 |
0 |
8 |
4 |
0 |
0 |
State Hospital, Iwo
(7.66686oN; 4.19926oE) |
Trauma |
22 |
10 |
4 |
6 |
3 |
0 |
3 |
1 |
1 |
1 |
0 |
0 |
| Skin lesions/abrasions |
4 |
2 |
0 |
2 |
2 |
0 |
2 |
1 |
1 |
0 |
0 |
0 |
| Caesarian section |
03 |
03 |
0 |
03 |
3 |
0 |
3 |
0 |
3 |
0 |
0 |
0 |
| Urine |
84 |
42 |
8 |
34 |
18 |
4 |
14 |
0 |
15 |
3 |
0 |
0 |
| Rectal swabs |
20 |
10 |
6 |
4 |
6 |
0 |
6 |
0 |
4 |
2 |
0 |
0 |
| Sub Total |
133 |
67 |
18 |
49 |
32 |
4 |
28 |
2 |
24 |
6 |
0 |
0 |
| Gross Total |
431 |
208 |
76 |
132 |
85 |
24 |
61 |
13 |
56 |
15 |
1 |
0 |
Altogether, 208 (48.3%) enterococci strains comprising of 85 (40.9%) VRE were recovered from 431 samples. The predominant Enterococcus specie was E. faecium: 88.5% (184/208) of recovered isolates were E. faecium (87 from State Specialist Hospital, Osogbo, 60 from State Hospital, Iwo, and 37 from Oke-Baale Primary Health Centre, Osogbo), while 24 (11.5%) were E. faecalis. However, 71.8% (61/85) of VRE were E. faecium (27 from State Specialist Hospital, Osogbo, 25 from State Hospital, Iwo, and 9 from Oke-Baale Primary Health Centre, Osogbo), while 28.2% (24/85) were E. faecalis (14 from State Specialist Hospital, Osogbo, 7 from State Hospital, Iwo and 3 from Oke-Baale Primary Health Centre, Osogbo). The highest percentage of VRE isolates was obtained from skin lesions and abrasions at 42.9% (12/28), followed by wounds from automobile accidents (trauma) at 23.0% (17/74). This was closely followed by CS wounds at 22.2% (4/18), while urine (45/269 samples) and rectal (7/42) samples had a recovery rate of 16.7% each. However, urine samples had the highest proportion of VRE strains at 52.9% (n = 45/85). The highest number of VRE isolates were recovered from participants within the age bracket 21 – 40 years, with urine samples having the highest rate of recovery of VRE at 53.6% (30/56 isolates) for that category (Table 2).
Antibiotic Resistance Profile of Recovered VRE Strains
All the screened isolates exhibited resistance to ampicillin (100%). The resistance was also high to clindamycin and Q/D at 97.6% and 92.9% respectively. None of the antibiotics exhibited 100% activity against all the isolates; and the highest susceptibility rate was observed with tigecycline at 72.9%, and was closely followed by nitrofurantoin at 71.8% (Figure 1). E. faecium from the three study locations exhibited higher resistance rates to all the antibiotics than E. faecalis, and this was evident for all the antibiotics. All isolates from the three selected hospitals were multidrug-resistant. The details of the resistance patterns of VRE isolates by species and study locations are shown in Table 3.
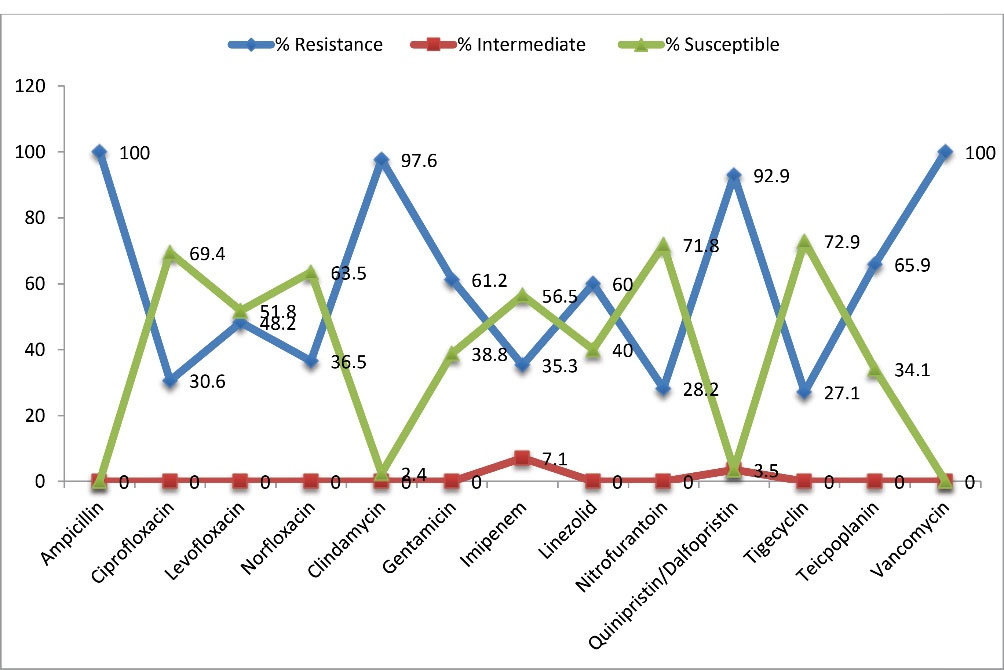
Figure 1.
Percentage Resistance/Susceptibility of the VRE Isolates to Various Antibiotics.
.
Percentage Resistance/Susceptibility of the VRE Isolates to Various Antibiotics.
Table 3.
Comparative Analysis of the Resistant Profiles of VRE Species by Study Location
|
Antibiotic class
|
Antibiotics
|
Total
a
|
Total
b
(%)
|
Frequency of Occurrence Of Resistant Isolates (%)
|
State Specialist Hospital, Osogbo
(n = 41)
|
Oke-Baale Primary Health Centre (n = 12)
|
State Hospital, Iwo (n = 32)
|
E. faecium
(n = 27)
|
E. faecalis
(n = 14)
|
E. faecium
(n = 9)
|
E. faecalis
(n = 3)
|
E. faecium
(n = 25)
|
E. faecalis
(n = 7)
|
| B-Lactams |
Ampicillin |
85 |
85 (100) |
27 (31.8) |
14 (16.5) |
9 (10.6) |
3 (3.5) |
25 (29.4) |
7 (8.2) |
| Fluoroquinolone |
Ciprofloxacin |
85 |
26 (30.6) |
7 (26.9) |
4 (15.4) |
4 (15.4) |
0 (0.0) |
7 (26.9) |
4 (15.4) |
| Levofloxacin |
85 |
41 (48.2) |
11 (26.8) |
5 (12.2) |
6 (14.6) |
2 (4.9) |
11 (26.8) |
6 (14.6) |
| Norfloxacin |
85 |
31 (36.5) |
8 (25.8) |
4 (12.9) |
6 (19.4) |
0 (0.0) |
9 (29.0) |
4 (12.9) |
| Lincosamides |
Clindamycin |
85 |
83 (97.6) |
26 (31.3) |
13 (15.7) |
9 (10.8) |
3 (3.6) |
25 (30.1) |
7 (8.4) |
| Aminoglycosides |
Gentamicin |
85 |
52 (61.2) |
15 (28.8) |
6 (11.5) |
6 (11.5) |
2 (3.8) |
16 (30.8) |
7 (13.5) |
| Carbapenems |
Imipenem |
85 |
30 (35.3) |
14 (46.7) |
3 (10.0) |
3 (10.0) |
0 (0.0) |
9 (30.0) |
1 (3.3) |
| Oxazolidinones |
Linezolid |
85 |
51 (60.0) |
20 (39.2) |
12 (23.5) |
6 (11.8) |
1 (2.0) |
11 (21.6) |
1 (2.0) |
| Nitrofurans |
Nitrofurantoin |
85 |
24 (28.2) |
9 (37.5) |
6 (25.0) |
2 (8.3) |
1 (4.2) |
6 (25.0) |
0 (0.0) |
| Streptogramin |
Quinupristin
/Dalfopristin |
85 |
79 (92.9) |
23 (29.1) |
13 (16.5) |
9 (11.4) |
3 (3.8) |
24 (30.4) |
7 (8.9) |
| Glycylcycline |
Tigecycline |
85 |
23 (27.1) |
9 (39.1) |
4 (17.4) |
3 (13.0) |
1 (4.3) |
6 (26.1) |
0 (0.0) |
| Glycopeptides |
Teicoplanin |
85 |
56 (65.9) |
20 (35.7) |
10 (17.9) |
5 (8.9) |
3 (5.4) |
15 (26.8) |
3 (5.4) |
| Vancomycin |
85 |
85 (100) |
27 (31.8) |
14 (16.5) |
9 (10.6) |
3 (3.5) |
25 (29.4) |
7 (8.2) |
Legend: Ampicillin (AMP), Ciprofloxacin (CIP), Clindamycin (DA), Gentamicin (CN), Imipenem (IMP), Levofloxacin (LEV), Linezolid (LZD), Nitrofurantoin (F), Norfloxacin (NOR), Quinupristin-Dalfopristin (Q/D), Teicoplanin (TEC), Tigecycline (TGC), Vancomycin (VAN). R = Resistance, S = Susceptible and I = intermediate.
One VRE strain displayed resistance to all the ten classes of antibiotics. This isolate withvanAphenotype (E. faecium) was recovered from the State Specialist Hospital, Osogbo from the urine sample of a 35-year-old female patient. Thirty-four VRE isolates expressed resistance to seven distinct classes of antibiotics (40.0%), alongside an additional 15 isolates resistant to 8 classes and above, making up 57.6% of the isolates resistant to 7 classes or more. Twenty-three isolates were resistant to 6 classes. The least resistance was to 3 classes of antibiotics again by only one isolate from the same hospital, this time vanB E. faecalis from the wound sample of a 46-year-old female.
All the VRE isolates had MAR indices ≥ 0.2, with 30.6% having MAR indices of 0.6. MAR indices were >0.2 in 97.6% of isolates from State Specialist Hospital, Osogbo, and 100.0% from both Oke-Baale Primary Health Centre and State Hospital, Iwo (Figure 2).
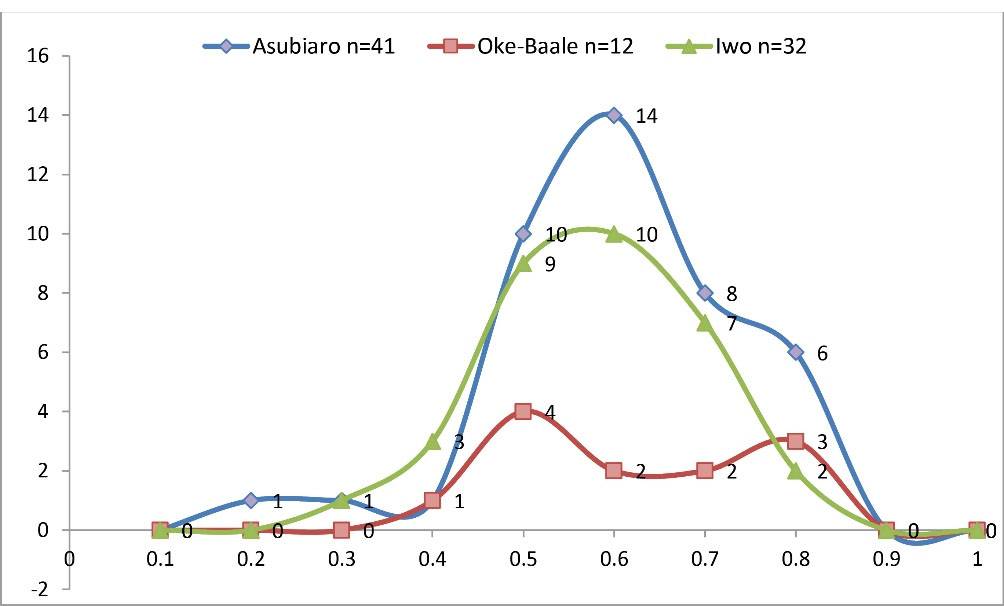
Figure 2.
Pattern of MAR Indices of VRE Isolates from Various Samples Recovered from Three Selected Hospitals
.
Pattern of MAR Indices of VRE Isolates from Various Samples Recovered from Three Selected Hospitals
Screening for vanAand vanB resistance phenotype was done by evaluating their susceptibility patterns to vancomycin and teicoplanin. Dual resistance to vancomycin and teicoplanin indicates the possible incidence of thevanAresistance gene, while resistance to only vancomycin is indicative of vanB resistance phenotype. The vanA resistant phenotype was higher at 65.9% (56/85) than the vanB resistant phenotype with a rate of 34.1% (29/85). The breakdown of the occurrence of van phenotypes of E. faecium and E. faecalis is depicted in Figure 3.
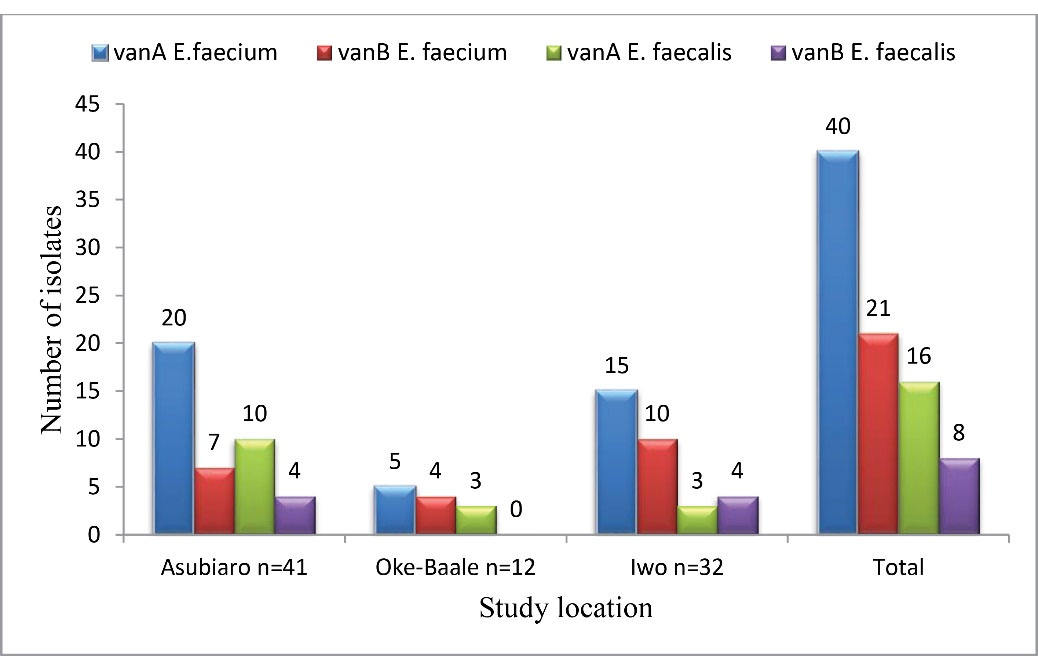
Figure 3.
Distribution of VRE Isolates by Phenotype in the Selected Hospitals
.
Distribution of VRE Isolates by Phenotype in the Selected Hospitals
Discussion
Enterococci are found in both the human and animal gastrointestinal tract, and as such is a constituent of the natural gastrointestinal microbiome. As they are proficient at surviving under a varied range of severe situations, they often cause opportunistic infections in the health-care setting. Predominant in human infections are E. faecium and E. faecalis.
The number of female participants was higher, probably because a high number of urine samples were collected from females at the antenatal clinic, a gender-specific activity as it involves only females. The State Specialist Hospital, Osogbo had the highest number of participants as it was the largest amongst the hospitals selected. It is a tertiary hospital established by the State Government which caters to the inhabitants of Osogbo and serves as a referral center to nearby towns. This is however at variance with the study of Moosavian et al (17) as most participants (68.0%) in their study were male.
Enterococcal infections are of immense importance in worldwide health challenges, resulting in extensive illness in the populace. The prevalence of enterococci in this study was 48.3%, and E. faecium was the major isolated specie. This rate correlates with that of another study in India (18), wherein E. faecium was also the major recovered specie. In this study, however, the percentage of enterococci colonization was lower than that of a study from Ethiopia (19) with a prevalence of 63%. This disparity could be due to the fact that samples in their study were recovered from HIV patients.
Various infections caused by VRE including urinary tract, wound, and bloodstream infections are on the increase in Nigeria (20). Prevalence rates of VREobtained from wound swabs, urine and rectal swabs in different studies vary drastically. This study reports a VRE prevalence rate of 40.9% (85/208) of which E. faecium was 71.8% (61/85) and E. faecalis 28.2% (24/85). These rates agree with the reports of other authors from different parts of the world where similar trends have been recorded (17). However, in Africa and in other parts of the world, different prevalence rates have been recorded in clinical samples vacillating between 5.7% to 88.9% (21). In similar studies from other authors, nevertheless, lower values of prevalence were recorded for E. faecium and higher prevalence for E. faecalis (22,23). These differences in prevalence rates are most likely due to variations in geographic locations. The prevalence rate recorded by our study is remarkably high enough to be worrisome. Previous studies have reported VRE transmission from the hands of caregivers in hospital wards (13,14), admission into ward spaces hitherto inhabited by patients colonized with VRE (24), contaminated surfaces of hospital wards such as floors, doorknobs, hand-rails, blood pressure cuffs, and hospital gowns (25), any of which factors is a possible means of transmission in all three hospitals included in the present study.
Reports of studies where higher prevalence rates among females than their male counterparts were recorded abound (19,21,26). This corroborates the present study as the infection rate of VRE in females was higher than that of males with a rate of 46.2% (61/132), while the VRE infection rate of males was 31.6% (24/76). However, this study revealed that infection by enterococci in males from the three selected study hospitals was essentially higher than that of females even though we had a higher number of female participants recorded during sample collection as 58.5% of the males were infected (76/130), while enterococcal infection rate in the female was 43.9% (132/301).
It is worthy of note that the major reasons that enterococci continues to exist in hospital settings are their ability to survive in diverse environments, their intrinsic ability to resist certain antibacterial agents, coupled with the development of diverse resistance patterns to many antibiotics (17) probably due to mutation or horizontal resistant-gene transfer. Within the last ten years, acquired resistance to aminoglycosides and glycopeptides, specifically vancomycin and teicoplanin employed in the therapy infections caused by enterococci, has become rampant (17). VanAand vanBphenotypes are most commonly detected in clinical VRE strains. In our study, the vanA phenotype was found to be higher at 65.9% (56/85) than the vanB phenotype (34.1%). Praharaj et al (27) reported that the majority of the VRE isolates (96.8%) in their study had the vanAphenotype, 3.3% of VRE isolates with vanC, while the vanBphenotype was not identified in any VRE strain. Even though we report only phenotypes in the present study, it has been reported that the vanBgeneis clustered and takes up a significant portion of the chromosome, much unlike thevanA gene. As such, it is less likely to be spread among strains. Moreover, thevanBgene is largely associated with epidemics and food contamination, while thevanAgene is linked to hospital isolates (3).
Antibiotic sensitivity test results in our study revealed that none of the antibiotics exhibited 100% activity against all the isolates. The most effective drugs in-vitro are tigecycline (72.9%), nitrofurantoin (71.8%), ciprofloxacin (69.4%), and norfloxacin (63.5%). This finding is at variance with a study (28) where isolates showed a much higher rate of sensitivity to nitrofurantoin at 93.0%. The least resistance in this study was observed with tigecycline; a tetracycline at 27.1%. The reasons for this pattern are not quite clear but increased production of tetracycline resistance factors tet(L)-encoded MFS (Major Facilitator Superfamily) pump and tet(M)-encoded ribosomal protection protein have been reported to be proficient at bestowing tigecycline resistance to clinical isolates of enterococci (29).
Studies conducted in Nigeria (20) and Iran (28) revealed 100% susceptibility of isolates to linezolid in contrast with our finding as 40% of our isolates were susceptible to linezolid. This could probably be a consequence of prior exposure to the drug, as 47.8% of our study population had been previously exposed to antibiotics (10). Other postulated reasons for the variances in outcomes could be the result of discrepancies in the study population, sample size, types of specimens, different procedures administered during isolation, level of hygiene of patients, environmental factors, regional differences, as well as contact with hospital settings.
In this study, all the recovered isolates were ampicillin resistant and multiply drug-resistant (100%), corresponding with a previous study (26). A high resistance rate to clindamycin (97.6%) was recorded in our study, a finding correlating with a study (30) that reported that >92% of E. faecium isolates had 96.0% resistance to clindamycin, while 100% resistance was recorded for the E. faecalis isolates. This is most likely due to the intrinsic resistance of enterococci generally to lincosamides (31).
Resistance to Q/D was also high at 92.9%. This is at extreme variance with an earlier report by Wanget al(32) with an incredibly low resistance rate of 1.0% to Q/D, although higher rates of resistance of enterococci to streptogramin (Q/D) was reported in Korea where resistance rates to Q/D by E. faecium was 10.0% and E. faecalis was 81.5% (33). As far as we know, there have been no reports of resistance of enterococci species and more specifically, VRE isolates to Q/D in Southwest Nigeria, and so, our study presents novel data in that regard. These high values of Q/D resistance in Southwestern Nigeria are worrisome as the drug is not frequently prescribed by clinicians. The resistance to Q/D was observed prior to its commercial usage use in the United States, suggesting that the appearance of the phenomenon may be attributed to other factors (33), and not necessarily associated with exposure to the drug (32). The resistance of enterococci species to Q/D has been reported to relate to enzymatic acetylation, efflux of the drug, and di-methylation of the 23S rRNA target site (34).
Higher levels of multidrug resistance were observed in E. faecium than in E. faecalis from all the study locations. This corresponds with various studies by different authors (17,22,23) as higher values of multidrug resistance in E. faecium were also reported. Multidrug-resistant strains of enterococci constitute serious problems in treatment regimens in patients with enterococcal infections (28). A previous study (35) reported that the percentage of VRE rose regarding resistance to a higher number of antibiotic classes, as 16%, 33%, and 100% of VRE isolates were resistant to 3, 4, and 5 antibiotic classes, respectively. While E. faecalis has been reported to be more implicated in human infections as a result of increased virulence, resistance to vancomycin is more associated with E. faecium isolates as it frequently exhibits multi-resistance characteristics (5) which are now progressively more credited to human infections such as bacteremia, endocarditis, urinary tract, and surgical site infections (4). This involvement may well be justified by their intrinsic and acquired resistance to several antibiotics together with great genomic variability, higher flexibility in the clinical environment, along incidence of a variety of virulence factors (4).
The high prevalence rate, as well as the elevated level of multidrug resistance, detected in our studyis worrisome and continues to be a threat in the therapy of infection elicited by these strains as vancomycin was identified as a last line of defense against enterococcal infections.Proper regulation guiding the use of antimicrobials in medical practice in addition to resolute controlof indiscriminate usage of antibiotics – oftentimes without prescriptions, must be ensured to eliminate selective pressure.
Conclusion
Herein, our study reportsthat the prevalence of VRE. faecium andVRE. faecalis in three selected hospitals in Southwest Nigeria was higher in females than in males. The antibiotic sensitivity pattern also revealed in-vitro effectiveness of tigecycline and nitrofurantoin against recovered VRE isolates in our study but records an excessive resistance rate to Q/D, a novel report in this region. All isolates were multidrug-resistant; this poses a great risk, as infections resulting from these organisms may complicate therapy and culminate in increased morbidity, as well as mortality.
Author Contributions
FMA designed, and supervised the research, participated in the bacteriological and molecular study, took part in data verification, interpretation, and analyses, and prepared the manuscript; N-AY contributed to study design, collected samples, performed the bacteriological and molecular investigation as well as other laboratory procedures, contributed to data interpretation. RRA and OOO participated in the bacteriological study and other laboratory procedures; KAA-N: analyzed and interpreted the data, contributed to manuscript preparation, and did the final proofreading. All authors read and approved the final version of the manuscript.
Conflict of Interests
The authors have declared that there are no competing interests.
Ethical Approval
Ethical approval for the study covering the three selected hospitals was gotten from the Health Research Ethics Board of the State Specialist Hospital, Osogbo (approval number - HREC/SSHO/11/478).
Funding
No funding was gotten for this study
Acknowledgments
The authors acknowledge with gratitude the technical staff of the Hospital Laboratories for their assistance, and participating patients for cooperating during sample collection.
References
- Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther 2014; 12(10):1221-36. doi: 10.1586/14787210.2014.956092 [Crossref] [ Google Scholar]
- Kolonen A, Sinisalo M, Huttunen R, Syrjänen J, Aittoniemi J, Huhtala H. Bloodstream infections in acute myeloid leukemia patients treated according to the Finnish Leukemia Group AML-2003 protocol - a prospective nationwide study. Infect Dis (Lond) 2017; 49(11-12):799-808. doi: 10.1080/23744235.2017.1347814 [Crossref] [ Google Scholar]
- Marothi YA, Agnihotri H, Dubey D. Enterococcal resistance--an overview. Indian J Med Microbiol 2005; 23(4):214-9. [ Google Scholar]
- Soheili S, Ghafourian S, Sekawi Z, Neela V, Sadeghifard N, Ramli R. Wide distribution of virulence genes among Enterococcus faecium and Enterococcus faecalis clinical isolates. ScientificWorldJournal 2014; 2014:623174. doi: 10.1155/2014/623174 [Crossref] [ Google Scholar]
- O’Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect Drug Resist 2015; 8:217-30. doi: 10.2147/idr.s54125 [Crossref] [ Google Scholar]
- Flokas ME, Karageorgos SA, Detsis M, Alevizakos M, Mylonakis E. Vancomycin-resistant enterococci colonisation, risk factors and risk for infection among hospitalised paediatric patients: a systematic review and meta-analysis. Int J Antimicrob Agents 2017; 49(5):565-72. doi: 10.1016/j.ijantimicag.2017.01.008 [Crossref] [ Google Scholar]
- Corso AC, Gagetti PS, Rodríguez MM, Melano RG, Ceriana PG, Faccone DF. Molecular epidemiology of vancomycin-resistant Enterococcus faecium in Argentina. Int J Infect Dis 2007; 11(1):69-75. doi: 10.1016/j.ijid.2006.02.003 [Crossref] [ Google Scholar]
- Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D. Ready for a world without antibiotics? the Pensières antibiotic resistance call to action. Antimicrob Resist Infect Control 2012; 1(1):11. doi: 10.1186/2047-2994-1-11 [Crossref] [ Google Scholar]
- Werner G, Klare I, Fleige C, Geringer U, Witte W, Just HM. Vancomycin-resistant vanB-type Enterococcus faecium isolates expressing varying levels of vancomycin resistance and being highly prevalent among neonatal patients in a single ICU. Antimicrob Resist Infect Control 2012; 1(1):21. doi: 10.1186/2047-2994-1-21 [Crossref] [ Google Scholar]
- Ulrich N, Vonberg RP, Gastmeier P. Outbreaks caused by vancomycin-resistant Enterococcus faecium in hematology and oncology departments: a systematic review. Heliyon 2017; 3(12):e00473. doi: 10.1016/j.heliyon.2017.e00473 [Crossref] [ Google Scholar]
- Lee T, Pang S, Abraham S, Coombs GW. Antimicrobial-resistant CC17 Enterococcus faecium: the past, the present and the future. J Glob Antimicrob Resist 2019; 16:36-47. doi: 10.1016/j.jgar.2018.08.016 [Crossref] [ Google Scholar]
- Iregbu KC, Ogunsola FT, Odugbemi TO. Susceptibility profile of Enterococcus faecalis isolated at the Lagos University Teaching Hospital, Nigeria. Niger Postgrad Med J 2002; 9(3):125-8. [ Google Scholar]
- Olawale KO, Fadiora SO, Taiwo SS. Prevalence of hospital-acquired enterococci infections in two primary-care hospitals in Osogbo, Southwestern Nigeria. Afr J Infect Dis 2011; 5(2):40-6. doi: 10.4314/ajid.v5i2.66513 [Crossref] [ Google Scholar]
- David OM, Oluduro AO, Olawale AK, Osuntoyinbo RT, Olowe OA, Famurewa O. Incidence of multiple antibiotic resistance and plasmid carriage among Enterococcus faecalis isolated from the hands of health care workers in selected hospitals in Ekiti, Ondo and Osun States, Nigeria. Int J Acad Res 2010; 2(1):43-7. [ Google Scholar]
- Quiloan MLG, Vu J, Carvalho J. Enterococcus faecalis can be distinguished from Enterococcus faecium via differential susceptibility to antibiotics and growth and fermentation characteristics on mannitol salt agar. Front Biol 2012; 7(2):167-77. doi: 10.1007/s11515-012-1183-5 [Crossref] [ Google Scholar]
- Krumperman PH. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl Environ Microbiol 1983; 46(1):165-70. doi: 10.1128/aem.46.1.165-170.1983 [Crossref] [ Google Scholar]
- Moosavian M, Ghadri H, Samli Z. Molecular detection of vanA and vanB genes among vancomycin-resistant enterococci in ICU-hospitalized patients in Ahvaz in southwest of Iran. Infect Drug Resist 2018; 11:2269-75. doi: 10.2147/idr.s177886 [Crossref] [ Google Scholar]
- Banerjee T, Anupurba S. Prevalence of virulence factors and drug resistance in clinical isolates of enterococci: a study from North India. J Pathog 2015; 2015:692612. doi: 10.1155/2015/692612 [Crossref] [ Google Scholar]
- Agegne M, Abera B, Derbie A, Yismaw G, Shiferaw MB. Magnitude of vancomycin-resistant enterococci (VRE) colonization among HIV-infected patients attending ART clinic in West Amhara government hospitals. Int J Microbiol 2018; 2018:7510157. doi: 10.1155/2018/7510157 [Crossref] [ Google Scholar]
- Ekuma AE, Oduyebo OO, Efunshile AM, Konig B. Surveillance for vancomycin resistant enterococci in a tertiary institution in South western Nigeria. Afr J Infect Dis 2016; 10(2):121-6. doi: 10.21010/ajid.v10i2.8 [Crossref] [ Google Scholar]
- Ali S, Alemayehu M, Dagnew M, Gebrecherkos T. Vancomycin-resistant enterococci and its associated risk factors among HIV-positive and-negative clients attending Dessie referral hospital, Northeast Ethiopia. Int J Microbiol 2018; 2018:4753460. doi: 10.1155/2018/4753460 [Crossref] [ Google Scholar]
- Bhat KG, Paul C, Ananthakrishna NC. Drug resistant enterococci in a south Indian hospital. Trop Doct 1998; 28(2):106-7. doi: 10.1177/004947559802800220 [Crossref] [ Google Scholar]
- Mendiratta DK, Kaur H, Deotale V, Thamke DC, Narang R, Narang P. Status of high level aminoglycoside resistant Enterococcus faecium and Enterococcus faecalis in a rural hospital of central India. Indian J Med Microbiol 2008; 26(4):369-71. doi: 10.4103/0255-0857.43582 [Crossref] [ Google Scholar]
- Drees M, Snydman DR, Schmid CH, Barefoot L, Hansjosten K, Vue PM. Prior environmental contamination increases the risk of acquisition of vancomycin-resistant enterococci. Clin Infect Dis 2008; 46(5):678-85. doi: 10.1086/527394 [Crossref] [ Google Scholar]
- Kramer A, Schwebke I, Kampf G. How long do nosocomial pathogens persist on inanimate surfaces? a systematic review. BMC Infect Dis 2006; 6:130. doi: 10.1186/1471-2334-6-130 [Crossref] [ Google Scholar]
- Abebe W, Endris M, Tiruneh M, Moges F. Prevalence of vancomycin resistant enterococci and associated risk factors among clients with and without HIV in Northwest Ethiopia: a cross-sectional study. BMC Public Health 2014; 14:185. doi: 10.1186/1471-2458-14-185 [Crossref] [ Google Scholar]
- Praharaj I, Sujatha S, Parija SC. Phenotypic & genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res 2013; 138(4):549-56. [ Google Scholar]
- Nasaj M, Mousavi SM, Hosseini SM, Arabestani MR. Prevalence of virulence factors and vancomycin-resistant genes among Enterococcus faecalis and E faecium isolated from clinical specimens. Iran J Public Health 2016; 45(6):806-13. [ Google Scholar]
- Fiedler S, Bender JK, Klare I, Halbedel S, Grohmann E, Szewzyk U. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J Antimicrob Chemother 2016; 71(4):871-81. doi: 10.1093/jac/dkv420 [Crossref] [ Google Scholar]
- Sattari-Maraji A, Jabalameli F, Node Farahani N, Beigverdi R, Emaneini M. Antimicrobial resistance pattern, virulence determinants and molecular analysis of Enterococcus faecium isolated from children infections in Iran. BMC Microbiol 2019; 19(1):156. doi: 10.1186/s12866-019-1539-y [Crossref] [ Google Scholar]
- Araújo TF, de Luces Fortes Ferreira CL. The genus Enterococcus as probiotic: safety concerns. Braz Arch Biol Technol 2013; 56(3):457-66. doi: 10.1590/s1516-89132013000300014 [Crossref] [ Google Scholar]
- Wang S, Guo Y, Lv J, Qi X, Li D, Chen Z. Characteristic of Enterococcus faecium clinical isolates with quinupristin/dalfopristin resistance in China. BMC Microbiol 2016; 16(1):246. doi: 10.1186/s12866-016-0863-8 [Crossref] [ Google Scholar]
- Oh WS, Ko KS, Song JH, Lee MY, Park S, Peck KR. High rate of resistance to quinupristin-dalfopristin in Enterococcus faecium clinical isolates from Korea. Antimicrob Agents Chemother 2005; 49(12):5176-8. doi: 10.1128/aac.49.12.5176-5178.2005 [Crossref] [ Google Scholar]
- Hershberger E, Donabedian S, Konstantinou K, Zervos MJ. Quinupristin-dalfopristin resistance in gram-positive bacteria: mechanism of resistance and epidemiology. Clin Infect Dis 2004; 38(1):92-8. doi: 10.1086/380125 [Crossref] [ Google Scholar]
- Arias CA, Murray BE. Enterococcus species, Streptococcus bovis group and Leuconostoc species. In: Mandell GL, Bennett JE, Dolin R, eds. Principles and Practice of Infectious Diseases. 7th ed. Philadelphia, PA: Churchill Livingstone, Elsevier; 2010. p. 2643-53.