Evaluation of the Frequency of Enterotoxin A (SEA) and Enterotoxin B (SEB) Genes in Clinical Isolates of Staphylococcus aureus in Rafsanjan, Iran
Avicenna J Clin Microbiol Infect, 6(4), 118-121; DOI:10.34172/ajcmi.2019.21
Original Article
Evaluation of the Frequency of Enterotoxin A (SEA) and Enterotoxin B (SEB) Genes in Clinical Isolates of Staphylococcus aureus in Rafsanjan, Iran
Afsaneh Mozafarianari1,2, Ashraf Kariminik2*, Mahnaz Tashakori3
1
School of Medicine, Immunology of Infectious Diseases Research Center, Rafsanjan University of Medical Sciences, Rafsanjan, Iran
2
Department of Microbiology, Kerman Branch, Islamic Azad University, Kerman, Iran
3
School of Allied Medical Sciences, Aliebn Abitaleb Educational and Treatment Hospital, Rafsanjan University of Medical Sciences, Rafsanjan, Iran
Corresponding author:
Ashraf Kariminik, Department
of Microbiology, Kerman
Branch, Islamic Azad
University, Kerman, Iran.
Tel: 00983431321769;
Cell phone:
00989133413556;
Email: a.kariminik@iauk.ac.ir
Abstract
Background: Staphylococcus aureus is one of the most important human pathogens that produces a wide range of toxins and causes various diseases. Staphylococcal enterotoxin is the most common cause of food poisoning. In addition, S. aureus enterotoxins are classified into 18 serotypes A to U based on serological and biological properties.
Methods: The samples were isolated from clinical specimens and identified by routine bacteriological methods. The isolated S. aureus was evaluated by polymerase chain reaction (PCR) for the detection of the genes encoding SEA and SEA.
Results: Based on the PCR results, 3 isolates possessed the enterotoxins B (SEB) gene while none of them showed enterotoxins A (SEA) gene.
Conclusions: The obtained results revealed that the clinical samples might be a potential source of the enterotoxigenic strains of S. aureus .
Keywords: Enterotoxin A, Enterotoxin B, Staphylococcus aureus, PCR
Background
Staphylococcus aureus is an important human pathogen, the virulence potential by which mainly relies on the production of an impressive set of protein toxins (1). This pathogen can work separately or in concert to cause a multitude of human diseases. Pneumonia, sepsis-related infections, toxic shock syndrome, and food poisoning are among the diseases that have traditionally and particularly been associated with the production of enterotoxins (2). In recent years, the prevalence of the bacterium is increasing in hospital-acquired infection and antibiotics prescribed to control and treat the infections have been ineffective due to the resistance phenomenon (3). However, there is no precise mechanism that can explain the invasive diseases caused by S. aureus (3), S. aureus species produce a variety of exoproteins such as enterotoxins (SEs), toxic shock syndrome toxin 1 (TSST1), exfoliative toxins, hemolysin coagulase, and leukocidin. About 15%-81% of S. aureus strains are able to produce enterotoxin (4). Enterotoxins are water-soluble proteins, which induce the non-specific T-cell proliferation. These toxins are stable to heat and proteolytic enzymes and could lead to food-borne disease (5). In addition, they stimulate the central nervous system via the toxin action on gastrointestinal nerve receptors and cause nausea, vomiting, and abdominal pain (6). Further, A and B enterotoxins are the most common and important exotoxins in food poisoning and hospital infection (7,8). Furthermore, enterotoxins A (SEA) is an important toxin in food poisoning and its gene is carried by the temperate bacteriophage (9). Moreover, enterotoxin B (SEB) is an agent in food poisoning and could lead to shortness of breath, widespread systemic damage, and even death in high doses. The SEB gene involves up to 900 nucleotides and is located on a chromosome (8). Various techniques are developed for the detection of relevant enterotoxins and genes. Polymerase chain reaction (PCR) is a simple, specific, reliable, and sensitive assay (7). PCR assay is used to determine the enterotoxin gene such as SEE, SEC, SEA and SEB. Using the molecular methods, the present study aimed to detect the enterotoxins A and B genes of S. aureus in the clinical samples of patients attending Abitaleb hospital in Rafsanjan, Iran.
Materials and Methods
Staphylococcus aureus Isolate Screening and Identification
In this descriptive, cross-sectional study, S. aureus isolates were randomly derived from patients who referred to Aliebn Abitaleb hospital, Rafsanjan, Iran in 2017. All samples were cultured in the blood agar and Mueller- Hinton agar media (Merck, Germany) and then incubated at 37° C for 48 hours. In general, 50 isolates of bacteria were screened after some diagnostic tests such as gram-staining, catalase, coagulase, DNase tests, and mannitol fermentation.
DNA Extraction and PCR
DNA was extracted by a DNA extraction kit (Sinaclon, Iran) following the manufacturer’s instructions. The quality and quantity of the extracted DNA were evaluated by spectrophotometry and 1% agarose gel electrophoresis. Additionally, PCR was performed for SEA and SEB genes. The applied primers were evaluated in the BLAST database to confirm the absence of nonspecific binding to the other regions of the genome. The sequences of the primers are shown in Table 1. The PCR reaction was accomplished according to the master mix (Sinaclon, Iran) protocol. The amplification reactions containing 1 μL of the forward primer, 1 μL of the reverse primer, 2 μL MgCl2 (50 mM), 0.5 μL dNTP, 2.5 μL buffers (10×), 2 μL of template DNA, 1 μL of Taq DNA polymerase, and 15 μL of distilled water. The S. aureus with SEA and SEB genes and Master Mix without DNA were considered as positive and negative controls, respectively. In addition, the PCR amplification was performed by the program on a Bio-Rad CFX96 system (Bio-Rad Laboratories Inc., Hercules, CA, USA) and the reaction was performed at 30 cycles (Table 2). The amplicons were analyzed by electrophoresis on 1.0% agarose gel, containing safe stain for 30-40 minutes at 90v, and finally, examined under the ultraviolet (UV) light using a UV light transilluminator (UV Star, Biometra, Germany).
|
Table 1. The Sequences of Primers
|
|
Gene
|
Primer
|
Sequences
|
Product Length
|
Reference
|
|
SEA
|
F
R |
5ʹ-TGTATGTATGGAGGTGTAAC-3ʹ
5ʹ-ATTAACCGAAGGTTCTGT-3ʹ |
270 bp |
(10) |
|
SEB
|
F
R |
5ʹ-TCGCATCAAACTGACAAACG-3ʹ
5ʹ-GCAGGTACTCTATAAGTGCC-3ʹ |
477 bp |
(10)
|
|
Note. SEA: Enterotoxin A; SEB: Entrotoxin B.
|
|
Table 2. Time and Temperature Used in PCR
|
|
Number
|
Steps
|
Temperature (°C)
|
Time
|
Number of Cycles
|
|
1
|
First denaturation |
94 |
30 seconds |
1 |
|
Denaturation |
94 |
4 minutes |
|
|
2
|
Annealing |
50 |
30 seconds |
30 |
|
Extension |
72 |
1 minute |
|
|
3
|
Final extension |
72 |
30 seconds |
1 |
|
Note. PCR: Polymerase chain reaction.
|
Results
After bacteriological diagnostic tests, all 50 isolates were gram-positive, beta-hemolytic, mannitol fermenting, along with catalase and coagulase positive. The SEA and SEB genes in the isolates of S. aureus were determined by the PCR method. The specificity of PCR was performed for positive and negative strains. The 7 SE -encoding genes (SEA and SEB) were detected in the positive strains but not in the negative strains (Figure 1). Furthermore, 270 and 477 bp segments were related to the amplification of a specific fragment of SEA and SEB genes that are responsible for enterotoxin type A and B and 270 bp for staphylococcal SEA gene (Figure 1, lanes 1 and 2), and 477 bp for staphylococcal SEB gene (Figure 1, lanes 4, 5, and 6). The result showed that 3 isolates (6%) of the clinical samples contained the SEB gene while none of the isolates contained the SEA gene (Figure 2).
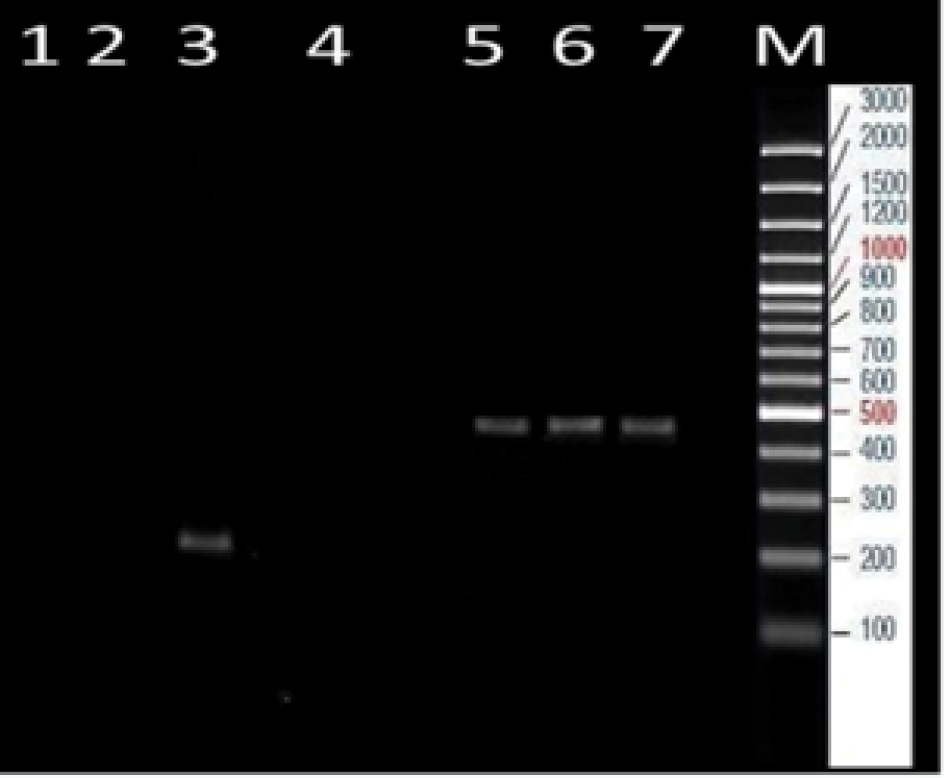
Gel Analysis of PCR-Amplified Toxin Gene Sequences.
Note. Lanes 1 and 2: SEA (270 bp); Lane 3: Positive control SEA ; Lanes 4, 5, and 6: SEB (477 bp); Lane 7: Negative control SEB ; M: Marker 100 bp.

The Distribution of S. aureus Isolates in Clinical Samples.
Discussion
Staphylococci are detectable on the skin and mucous membranes of humans (11). This bacterium might induce allergic inflammatory responses via secreting enterotoxin (SE) and could lead to toxic shock syndrome toxin 1 (TSST-1). Humans are the main storage for S. aureus in nature. Approximately 30% of healthy people are involved in a bacterium (7,12). In the present study, the presence of enterotoxin A and B was investigated in the clinical samples of patients in the hospital. A total of 6% of the samples contained SEB while none of them had SEA genes. A study was performed on the strains of S. aureus derived from patients who were in the hospital over 5 months and investigated the detection of enterotoxins using the real-time fluorescence PCR and reversed passive latex agglutination (SET-RPLA) methods. Based on the results, the PCR method was more suitable than SET-RPLA in the detection of enterotoxins gene. The results further demonstrated that PCR assay could detect some more enterotoxin genes while SET-RPLA was not able to detect them. Furthermore, it was found that the number of the SEA was less than SEB in the patient-derived S. aureus as shown by our study (7). Similarly, Fluer et al investigated the prevalence of S. aureus enterotoxins in the blood and burn wound of the patients by an indirect hemagglutination test and reported that 3% and 76%of the isolated strains were from burn wound and blood generate SEA, respectively (13). Moreover, Pourmand et al studied the prevalence of the SEA gene in the clinical sample of S. aureus and showed that 46.9% strains of S. aureus contained the SEA gene (14). The results of these studies are inconsistent with our results. Another researcher screened the S. aureus nasal strains to detect toxin genes and showed that 15% and 13% of the genes were TST and SEC, respectively. Methicillin-resistant S. aureus (MRSA) strains were detected in 4.5% of the students, indicating that the nasal was a perfect site for the growth of MRSA as in our study, bronchial samples contained enterotoxin genes (15). Other investigators defined the rate of enterotoxin gene-positive S. aureus isolates among the food handler in central Iran and explained that the rate of SEA was more than SEB in that food (16). Additionally, Arabestani et al screened S. aureus strains from a patient in the west of Iran. They demonstrated that the enterotoxin A gene had the most prevalence in the isolated strains. Their results showed that MRSA strains had the most resistance to cefoxitin and ciprofloxacin while they were completely sensitive to vancomycin (17). In another study, Pourmand et al reported that 98% and 97% of the isolates of SEA -positive and SEA -negative S. aureus were sensitive to vancomycin, respectively (14). Some studies evaluated the enterotoxigenicity of S. aureus in dairy products and concluded that these isolates contained a variety of SEs and demonstrated different antimicrobial resistance but almost all of them were resistant to vancomycin (11,18). The results of the present study showed that the prevalence of SE genes is different in a variety of strains as presented in our investigation, the frequency of SEB was 6% whereas that of SEA was not obvious in any strains.
Conclusions
The obtained results revealed that the clinical samples might be a potential source of the enterotoxigenic strains of S. aureus. These findings highlighted the need for strict hygienic and preventive measurements to avoid human health threats.
Conflict of Interest Disclosures
No competing interest was declared by any of the authors.
Acknowledgments
The authors of this article sincerely would like to thank Rafsanjan University of Medical Sciences and the Islamic Azad University of Kerman for financial support.
References
- do Vale A, Cabanes D, Sousa S. Bacterial toxins as pathogen weapons against phagocytes. Front Microbiol 2016;7:42. doi: 10.3389/fmicb.2016.00042. [Crossref]
- Fisher EL, Otto M, Cheung GYC. Basis of virulence in enterotoxin-mediated staphylococcal food poisoning. Front Microbiol 2018;9:436. doi: 10.3389/fmicb.2018.00436. [Crossref]
- Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 2011;6(4):e17936. doi: 10.1371/journal.pone.0017936. [Crossref]
- Argudín MA, Mendoza MC, González-Hevia MA, Bances M, Guerra B, Rodicio MR. Genotypes, exotoxin gene content, and antimicrobial resistance of Staphylococcus aureus strains recovered from foods and food handlers. Appl Environ Microbiol 2012;78(8):2930-5. doi: 10.1128/aem.07487-11. [Crossref]
- Otto M. Staphylococcus aureus toxins. Curr Opin Microbiol 2014;17:32-7. doi: 10.1016/j.mib.2013.11.004. [Crossref]
- Kurjogi M, Satapute P, Jogaiah S, Abdelrahman M, Daddam JR, Ramu V, et al. Computational modeling of the staphylococcal enterotoxins and their interaction with natural antitoxin compounds. Int J Mol Sci 2018;19(1). doi: 10.3390/ ijms19010133. [Crossref]
- Klotz M, Opper S, Heeg K, Zimmermann S. Detection of Staphylococcus aureus enterotoxins A to D by real-time fluorescence PCR assay. J Clin Microbiol 2003;41(10):4683-7. doi: 10.1128/jcm.41.10.4683-4687.2003. [Crossref]
- Liu PF, Wang Y, Ulrich RG, Simmons CW, VanderGheynst JS, Gallo RL, et al. Leaf-encapsulated vaccines: agroinfiltration and transient expression of the antigen staphylococcal endotoxin B in radish leaves. J Immunol Res 2018;2018:3710961. doi: 10.1155/2018/3710961. [Crossref]
- Balaban N, Rasooly A. Staphylococcal enterotoxins. Int J Food Microbiol 2000;61(1):1-10. doi: 10.1016/s0168- 1605(00)00377-9. [Crossref]
- Bokaeian M, Saeidi S, Hassanshahian M. Molecular detection of Staphylococcus aureus enterotoxin A and B genes in clinical samples from patients referred to health centers in Zahedan City. Res Mol Med 2016;4(2):44-6. doi: 10.18869/acadpub. rmm.4.2.44. [Crossref]
- Rahimi E. Enterotoxigenicity of Staphylococcus aureus isolated from traditional and commercial dairy products marketed in Iran. Braz J Microbiol 2013;44(2):393-9. doi: 10.1590/s1517- 83822013000200008. [Crossref]
- Ludwig S, Jimenez-Bush I, Brigham E, Bose S, Diette G, McCormack MC, et al. Analysis of home dust for Staphylococcus aureus and staphylococcal enterotoxin genes using quantitative PCR. Sci Total Environ 2017;581-582:750- 5. doi: 10.1016/j.scitotenv.2017.01.003. [Crossref]
- Fluer FS, Prokhorov V, Bondarenko VM, Dmitrienko OA, Lashenkova NN, Men’shikova ED, et al. [Isolation rate of enterotoxigenic staphylococci in patients with sepsis, pneumonia and burns]. Zh Mikrobiol Epidemiol Immunobiol. 2005(5):3-6.
- Pourmand MR, Memariani M, Hoseini M, Bagherzadeh Yazdchi S. High prevalence of sea gene among clinical isolates of Staphylococcus aureus in Tehran. Acta Med Iran. 2009:47(5):357-61.
- Piechowicz L, Garbacz K, Wiśniewska K, Dąbrowska-Szponar M. Screening of Staphylococcus aureus nasal strains isolated from medical students for toxin genes. Folia Microbiol (Praha) 2011;56(3):225-9. doi: 10.1007/s12223-011-0041-1. [Crossref]
- Fooladvand S, Sarmadian H, Habibi D, van Belkum A, Ghaznavi-Rad E. High prevalence of methicillin resistant and enterotoxin gene-positive Staphylococcus aureus among nasally colonized food handlers in central Iran. Eur J Clin Microbiol Infect Dis 2019;38(1):87-92. doi: 10.1007/s10096- 018-3398-0. [Crossref]
- Arabestani MR, Rastiyani S, Alikhani MY, Mousavi SF. The relationship between prevalence of antibiotics resistance and virulence factors genes of MRSA and MSSA strains isolated from clinical samples, West Iran. Oman Med J 2018;33(2):134-40. doi: 10.5001/omj.2018.25. [Crossref]
- Morandi S, Brasca M, Lodi R, Cremonesi P, Castiglioni B. Detection of classical enterotoxins and identification of enterotoxin genes in Staphylococcus aureus from milk and dairy products. Vet Microbiol 2007;124(1-2):66-72. doi: 10.1016/j.vetmic.2007.03.014. [Crossref]