Molecular Typing of ccrB Gene in Methicillin-resistant Staphylococcus aureus by Restriction Fragment Length Polymorphism
Avicenna J Clin Microbiol Infect, 6(4), 100-105; DOI:10.34172/ajcmi.2019.18
Original Article
Molecular Typing of ccrB Gene in Methicillin-resistant Staphylococcus aureus by Restriction Fragment Length Polymorphism
Fatemeh Noorbakhsh1*, Parvaneh Atayifar1, Shohreh Zare Karizi2
1
Department of Microbiology, Biological Science College, Varamin-Pishva Branch, Islamic Azad University, VaraminPishva, Iran
2
Department of Genetics, Biological Science College, Varamin-Pishva Branch, Islamic Azad University, Varamin-Pishva, Iran
*Corresponding author:
Fatemeh Noorbakhsh, Ph.D.
Department of Microbiology,
Islamic Azad University,
Varamin-Pishva Branch,
Varamin, Iran, P.O. BOX:
33817-74895,
Tel: 09122043654 Email: niloofar_noorbakhsh@yahoo.com
Abstract
Background: Staphylococcus aureus is one of the most important pathogens acquired from the hospital and community. Increasing the resistance of
S. aureus to antibiotics is a major health concentration, and thus the study of antibiotic resistance in
S. aureus is very important. The aim of this study is to determine the typing of methicillin-resistant
S. aureus (MRSA) in the region of the
ccrB gene by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP).
Methods: One hundred and six S. aureus were isolated from urine, blood, sputum, wound, and the trachea of patients hospitalized in Tehran during (March-April) 2016. Antibiotic susceptibility test was done by the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI). In addition, molecular typing for staphylococcal cassette chromosome mec (Sccmec) type I-V was performed in MRSA isolates, followed by conducting PCR-RFLP by restriction enzymes BsmI and HinfI in the ccrB gene area.
Results: PCR and typing showed that type II SCCmec was 40% (n=20), followed by types III (28, 56%), IVc (12, 24%), I (11, 22%), V (9, 18%) IVa (7, 14%), and IVb (5, 10%). However, SCCmec type IVd was not observed in the isolates. Finally, after the amplification of ccrB gene and RFLP, all isolates the same as the typing method represented types I, II, III, IVa, IVb, and IVc while no type V was detected by this method.
Conclusions: The results of this study demonstrated that SCCmec (type I-IV) can be detected by PCR-RFLP in the ccrB gene, but this method identified no type V SCCmec in MRSA
Keywords: Staphylococcus aureus, SCCmec, Typing, ccrB, PCR-RFLP
Background
Staphylococcus aureus is considered as one of the most essential bacterial pathogens in humans that can cause various infections in patients ranging from skin infections to fatal necrotizing pneumonia, bacteremia, and endocarditis (1). Methicillin-resistant S. aureus (MRSA) is one of the important pathogens that is responsible for many nosocomial infections. First, MRSA was identified only in the hospitals, but 30 years later, the first virulent MRSA was acquired from the community (2). The resistance to methicillin in S. aureus is induced by the presence of the mecA gene, encoding low-affinity penicillin-binding protein PBP2a (78 KD) (3,4). MRSA has a mobile genetic element staphylococcal cassette chromosome mec (SCCmec) carrying the mecA gene. (5-7) SCCmec elements in S. aureus are as unique genomic islands with 2 essential components (i.e., the ccr and the mec gene complexes) and J region (5,8,9). The ccr gene complex is composed of ccr genes encoding 2 site-specific recombinases (ccrA and ccrB), and the mecA gene complex contains mecA and regulatory genes mecI and mecR . ( 6,10,11) Zhang et al defined 8 different types of SCCmec in the combination of ccr and mec complex. While types I-V were widespread (12), other types existed in the strains of the country from which they were originated (13,14). SCCmec exchange between species is related to the ccr gene expression (15). Several allotypes of ccr and mec gene are classified in SCCmec . The 5 allotypes of the ccr gene complex include ccrAB1, ccrAB2, ccrAB3, ccrAB4, and ccrC (16,17), and 5 classes of the mec gene complex (types I-V) were described and SCCmec type IV has 8 individual subtypes (18,19).
The site-specific recombination of SCCmec is catalyzed by its encoded ccr recombinases, ccrA and ccrB for types I to IV and ccrC for type V. In addition, ccrA and ccrB belong to a family of large serine invertase and resolvases
which consist of resolvases, invertases, phage integrases, and transposases (6,20).
Further, ccrB gene as a target gene is often chosen because the sequence is highly conserved compared to the ccrA gene (21).
Molecular techniques for the typing of the used microbes include PFGF, methods based on restriction enzyme, the analysis of plasmid, and DNA typing method based on polymerase chain reaction (PCR) (22). Pulse-field gel electrophoresis and multi-locus sequence typing are the best techniques for phenotypic and genotypic studies, but the most important problems of this method are the technical complexity, high cost, as well as a longer process (23). Studies suggest that PCR restriction fragment length polymorphism (PCR-RFLP) can be replaced by these expensive and time-consuming techniques (22). Antibiogram is regarded as one of the most important typing methods in many hospitals since it is easily standardized (24).
Objectives
The aim of this study was the molecular typing of ccrB gene in MRSA by RFLP.
Materials and Methods
Bacterial Isolates
A total of 106 specimens doubtful to S. aureus were isolated from blood, urine, wound, nasal fluid, and the sputum of patients hospitalized in Milad hospital during March-April 2016. S. aureus isolates were identified based on gram-staining, catalase, coagulase test, and growth on the mannitol salt agar.
Antibiotic Susceptibility Test
The presence of the mecA gene and resistance to methicillin in S. aureus was confirmed by the oxacillin/ cefoxitin-resistant in all isolates. In addition, the antibiotic susceptibility test was performed by the disk diffusion method according to CLSI 2017. Further, the tested antibiotics were oxacillin (30 μg)⁄cefoxitin (30 μg) prepared from Padtan Teb. S. aureus ATCC 25923 was used as the control strain in the antibiotic susceptibility test.
DNA Isolation
MRSA isolates were cultured on brain heart infusion agar and incubated overnight in 37ºC. The bacteria were harvested from the medium and washed by the saline buffer. Then, DNA was extracted by the boiling method and the quality of the DNA was determined by the electerophores (25).
Amplification of SCCmec Type
Multiplex PCR for the detection of MRSA isolates SSCmec Type (I-V) were previously optimized for standard strains by Ito and Katayama (10). The primers of SCCmec types I–V including type IV subtypes (Table 1) were previously selected and reported (36). Further, the PCR mixture (Cinnagen) contained 12.5 μL of master mix (400 μm of dNTPs, 3 mM mgcl2, and 1.2 u Taq polymerase), 9.5 μL nuclease-free water, 10 pM each primer, and 1 μg DNA. Furthermore, the amplification for SCCmec types I, II, III, and V was performed by initial denaturation at 95ºC for 5 minutes, followed by 30-cycle denaturation at 95ºC for 30 seconds, annealing at 58ºC for 1 minute, and
extension at 72ºC for 2 minutes. Moreover, the multiplex PCR carried out for IV subtypes encompassed the initial denaturation at 95ºC for 5 minutes, followed by 30-cycle denaturation at 95ºC for 30 seconds, annealing at 59ºC for 30 seconds and extension at 72ºC for 2 minutes, as well as ending by final extension at 72ºC for 8 minutes and hold step at 5ºC. Finally, PCR products were electrophoresed in 1.5% gel agarose containing 1 μL safe stain and image supplied by UV transilluminator and gel document.
|
Table 1. The Profile of SCCmec Type Specific Primers
|
|
Primer
|
Orientation
|
Primer sequence
|
Target Gene
|
Size (bp)
|
| Type I |
Forward
Reverse |
GCTTTAAAGAGTGTCGTTACAGG
GTTCTCTCATAGTATGACGTCC |
ORF E008 of strain NCT C10442 |
613 |
| Type II |
Forward
Reverse |
GATTACTTCAGAACCAGGTCAT
TAAACTGTGTCACACGATCCAT |
kdpE of strain N315 |
287 |
| Type III |
Forward
Reverse |
CATTTGTGAAACACAGTACG
GTTATTGAGACTCCTAAAGC |
J1 region of SCCmec Type III |
243 |
| Type Iva |
Forward
Reverse |
GCCTTATTCGAAGAAACCG
CTACTCTTCTGAAAAGCGTCG |
ORF CQ002 of strain CA05 |
776 |
| Type IVb |
Forward
Reverse |
AGTACATTTTATCTTTGCGTA
AGTCATCTTCAATATGGAGAAAGTA |
J1 region of SCCmec type IVb |
994 |
| Type IVc |
Forward
Reverse |
TCTATTCAATCGTTCTCGTATT
TCGTTGTCATTTAATTCTGAACT |
IVc element of strain 81/108 |
677 |
| Type IVd |
Forward
Reverse |
AATTCACCCGTACCTGAGAA
AGAATGTGGTTATAAGATAGCTA |
CD002 in type IVd |
1242 |
| Type V |
Forward
Reverse |
GAACATTGTTACTTAAATGAGCG
TGAAAGTTGTACCCTTGACACC |
ORF V011 of strain JCSC3624 |
325 |
|
ccrB
|
Forward
Reverse |
GGCTATTATCAAGGCAATTTACC
ACTTTATCACTTTTGACTATTTCG |
|
643 |
Amplification of ccr B Gene
The primer sequence for the amplification of the ccrB gene was selected from the study by Yang et al (22) (Table 1). The total volume of PCR master mix was 50 μL containing 25 μL of master mix (400 μm of dNTPs, 3 mM mgcl2, and 1.2 u Taq polymerase), 20 μL nuclease-free water, 1.5 μL (10 pM) each primer, and 2 μL chromosomal DNA. Moreover, the cycling conditions included an initial step at 94ºC for four minutes, followed by 30 cycles of 94°C for 30 seconds, 59°C for 1 minute, 72°C for 2 minutes, and the final step 72°C for 8 minutes.
Polymerase Chain Reaction-Restriction Fragment Length Polymorphism
RFLP was used to detect the molecular typing of SCCmec (22). After the amplification of the ccrB gene in the MRSA strain, the PCR product was digested by HinfI and BsmI enzymes. Additionally, the digestion reaction was performed in two steps as follows.
Step 1 : PCR products were mixed with 17 μL nuclease-free water, one unit HinfI (Fermentas), and a two-unit buffer in a final reaction volume of 30 μL and then incubated at 37°C for 16 hours.
Step 2 : A total of 10 μL of step 1 product was mixed with 17 μL nuclease-free water, one unit BsmI (Fermentas), and a two-unit buffer in a final reaction volume of 30 μL and incubated at 37°C for 10 minutes, followed by adding 2 μL ethylenediaminetetraacetic acid (EDTA) (0.5 M) and incubating at 80°C for 20 minutes. The restriction fragments were separated by 2% agarose gel electrophoresis in Tris-acetate-EDTA buffer for 50 minutes at 90 V and stained by the ethidium bromide. Eventually, SPSS Statistics, version 22 was used to analyze the data.
Results
Bacterial Isolates
Staphylococcus aureus from the clinical samples in Milad hospital were isolated from different sources including (n=32, 30.1%) urine, (n=18, 17%) blood, (n=21, 19.8%) wound, (n=15, 14.15%) trachea, (n= 9, 8.5%) nasal fluid, and (n=11, 10.4%) sputum.
The average age of the evaluated patients was 45 years with a minimum of 15 and a maximum of 75 years. In addition, 45 and 61 strains were collected from men (42.45%) and women (57.54%), respectively.
Antibiotic Susceptibility Test
Antibiotic susceptibility of 106 strains of S. aureus to cefoxitin/oxacillin showed that 50 (47.1%), 36 (34%), and 20 (18.9%) isolates were resistant, susceptible, and intermediate, respectively. Fifty strains as MRSA were used for the molecular typing of SCCmec.
SCCmec Typing
A clear discriminated band pattern was obtained for all five types and subtypes of the main SCCmec using the PCR (Figure 1) and multiplex PCR (Figure 2). The individual PCR band size of each fragment for SCCmec types I, II, III, V, IVa, IVb, IVc, and IVd were 613, 287, 200, 325, 776, 994, 677, and 1242 bp, respectively. All the MRSA strains were positive in a certain SSCmec type. The most common type was type III (56%) while IVd subtype was not detected in this study (Figure 3).
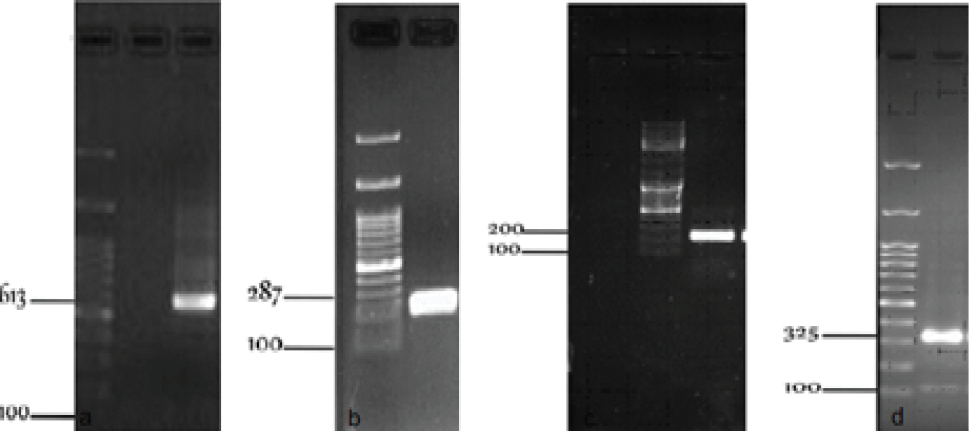
The Amplification of S. aureus SSCmec Type: (a) Type I, (b) Type II, (c) Type III, and (d) Type V.
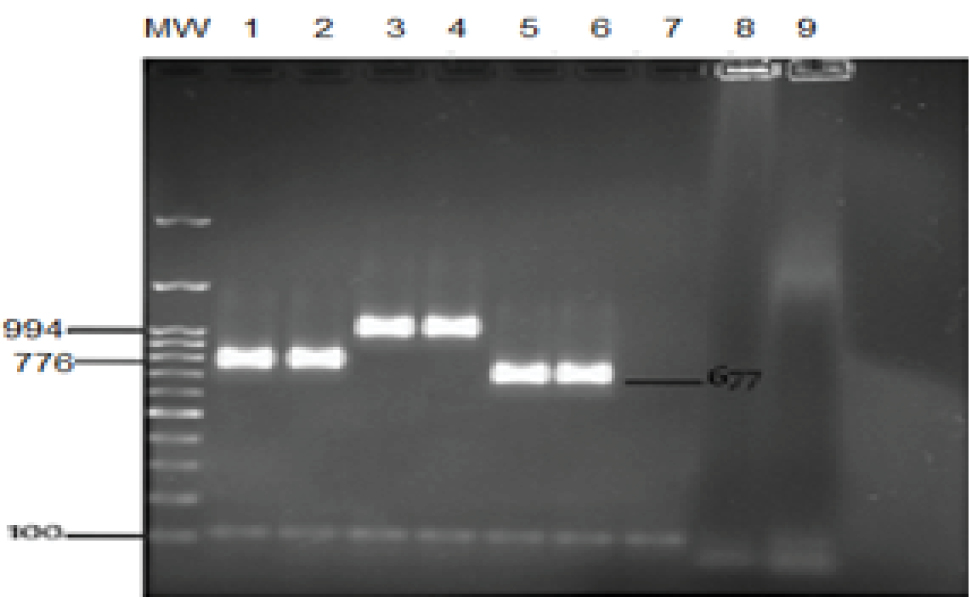
Type IV: MW: Marker 3kb + 100 bp, 1, 2: Type IVa; 3, 4: Type IVb; 5, 6: Type IVc; 7, 8: IVd, 9: Negative control.
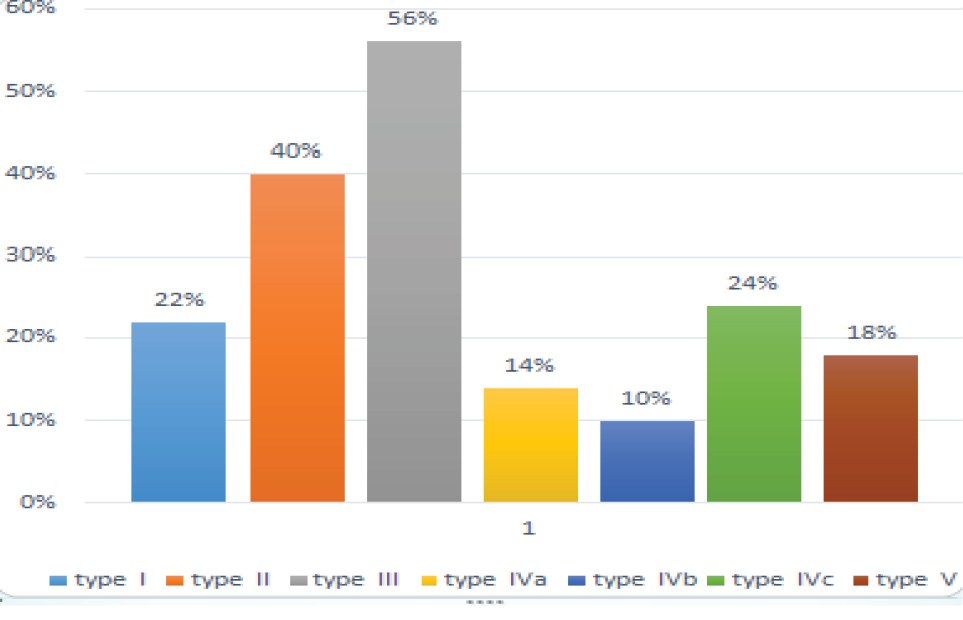
The Frequency of SSCmec Type and Subtype.
Rapid Typing
After the amplification of the ccrB gene, the PCR product was used to digest the two steps of PCR-RFLP (22). SCCmec typing method was applied for 50 strains of MRSA isolates. The results in electrophoresis were shown as type I with 1 band of 404 bp, type II with 1 band of 530 bp, type III with 2 bands of 218 and 225 bp, type Iva with 2 bands of 311 and 200 bp, type IVb with 2 bands of 530 and 600 bp, as well as type IVc with 3 bands of 311, 530, and 600 bp (Figure 4). Type IVd established 3 bands of 154, 225, and 264 bp, but type IVd was not detected in this study. This protocol is a suitable method for the rapid typing of the SCCmec element types I to IV, but the PCR-RFLP method is not likely to identify type V.
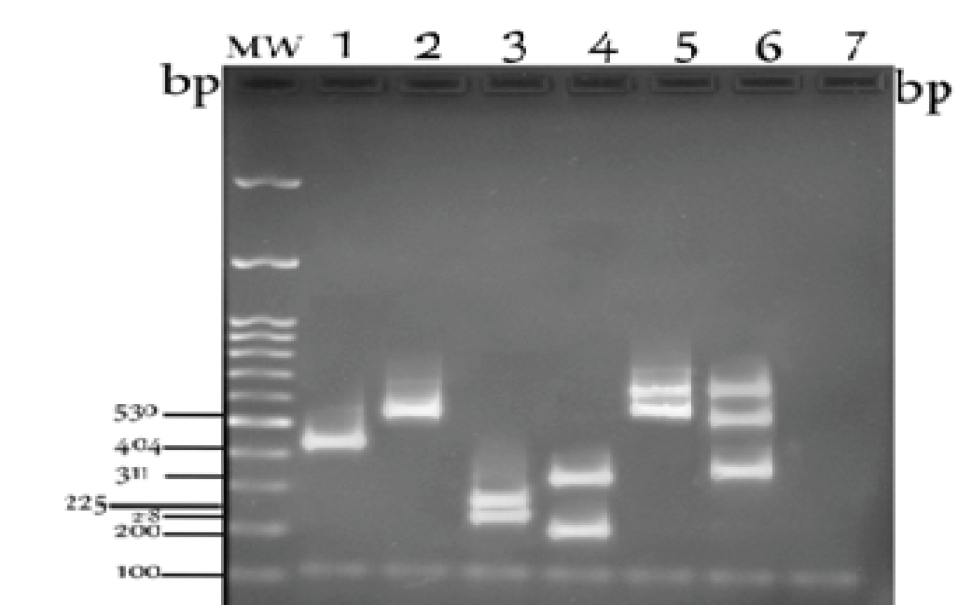
Agarose Gel Electrophoresis Showing 6 PCR-RFLP Patterns of Types I to IV Digested by HinfI (one step), as well as HinfI and BsmI (two steps).
Discussion
Methicillin-resistant (MRSA) gene in S. aureus is coded by chromosomal cassette mec and contains five main types of SCCmec. These organisms cause severe rates of disease and mortality worldwide. In the present study, 100 samples of S. aureus strains were isolated from hospitalized patients. The antibiotic susceptibility test revealed that 50 strains were resisted to oxacillin/cefoxitin. Further, methicillin resistance in staphylococci isolated from clinical samples by Rahbar was 53% in Tehran and 51% of MRSA were also reported in Turkey (26).
In this study, SCCmec typing was performed on 50 MRSA strains and all strains were typeable encompassing types III (28 cases), II (20), IVc (12), I (11), V (14), IVa (7), IVb (5), and IVd (0). Types III and IV were identified as the dominant types in this study, which mainly cause multi-drug resistant leading to increased health problems, especially in the hospital. Furthermore, types III and II exist together in 20% of MRSA strains and types II, III, IVb, and V were present together only in one bacterium although SCCmec type was observed in none of the cases 4, 5, 6,... Four subtypes of SCCmec type IV were not detected together in any strains.
Amiri et al evaluated 3 type I SCCmec, 12 type II SCCmec, 8 type IVb SCCmec, 4 type IVd SCCmec, and 3 type V SCCmec isolates. The findings revealed different types of SCCmec carries in MRSA strains in a hospital in Kashan and two dominant types in this study were types II and IV SCCmec (27). In another study, Abdollahi et al identified that 15 cases related to types I, IV, and V and 63 cases belonged to types II and III. Moreover, the highest type related to different types of II (34 cases), III (29 cases), and other types were I (6 cases), V (six), and IV (three). (28) The typing of MRSA using M-PCR included types III (33.33%), IV (43.33%), and V (23.33%), (12), which is somewhat similar to our study. Several studies indicated that SCCmec type III was to be circulating in Iranian hospitals and other Asian countries (29-31) and SCCmec type IV was the most frequent isolation of MRSA in healthy carries (32) whereas SCCmec type V was dominant in methicillin-resistant S. haemolyticus (30) in Iran.
In Slovenia, from 31 strains of MRSA, 16 isolates (51.6%) of SCCmec type IV, 7 (22.5%) and 2 (5.6%) strains were of SCCmec types I and III, respectively, and 6 strains (19.4%) were classified as non-typed. In Malaysia, among the 66 observed strains of MRSA, 52 cases (78.8%) were of SCCmec type III and 12 cases (18.18%) belonged to SCCmec type II (33). Chongtrakool et al showed that SCCmec type III was the most common type of mecA in 8 Asian countries (34). These differences in the literature indicate that different locations and the variety of treatment may affect the epidemic distribution of SCCmec type in the world.
The distribution of SCCmec in nature is limited to relatively few clonal complexes of related MRSA (35). The majority of epidemic H-MRSAs carries SCCmec types I, II or III (18). SCCmec subtype IVa or IVb carries non-oxacillin-resistant S. aureus strains (36), but SCCmec subtype IVc was observed in the hospital-acquired strains (37). Additionally, SCCmec IV is present in diverse genetic backgrounds, which suggests that type IV is a mobile element (38).
In this study, rapid SCCmec typing was investigated by the PCR-RFLP for the ccrB gene using two Hinfl and BsmI enzymes in MRSA, which matched the results obtained by the PCR assay. Similarly, SCCmec types II and IV were identified by one-step digestion with HinfI. In addition, 24 (12 IVc, 7 IVa, and 5 IVb subtypes, respectively) isolates of SCCmec type IV and 20 isolates of SCCmec type II were successfully discriminated by one-step digestion. The second step with BsmI digestion is necessary for discriminating of SCCmec type I and III. In general, 28 isolates of SCCmec type III and 11 isolates of SCCmec type I were detected in the second step.
Zhang et al detected SCCmec types IV, II, and III by the PCR-RLFP method. However, no SCCmec type I strains was found among the clinical MRSA isolates in this study (14). Type V cannot be identified by the PCR-RFLP method since the ccrB gene of types I to IV has 37.4% homology to the ccrC gene of type V at the nucleic acid level (39,40).
Conclusions
The findings revealed that using the PCR-RFLP method can be used to identify different types of staphylococcal cassette chromosome mec (SCCmec). This method can replace the original method with eight pairs of primers for SCCmec typing whereas the PCR-RFLP method is only performed with one pair of primers and one or two restriction enzymes.
Ethical Approval
Not applicable.
Conflict of Interest Disclosures
The authors declare that there is no financial or commercial conflict of interest in this study.
Acknowledgments
The authors would like to thank the microbiology laboratory personnel of Islamic Azad University, Pishva Branch for their cooperation.
References
- Mahon CR, Lehman DC, Manuselis G Jr. Textbook of Diagnostic Microbiology. 3th ed. Saunders; 2007. p. 369.
- Turlej A, Hryniewicz W, Empel J. Staphylococcal cassette chromosome mec (SCCmec ) classification and typing methods: an overview. Pol J Microbiol 2011;60(2):95-103.
- Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett 1987;221(1):167-71. doi: 10.1016/0014-5793(87)80373-3. [Crossref]
- Namvar AE, Afshar M, Asghari B, Rastegar Lari A. Characterisation of SCCmec elements in methicillin-resistant Staphylococcus aureus isolated from burn patients. Burns 2014;40(4):708-12. doi: 10.1016/j.burns.2013.09.010. [Crossref]
- Ito T, Okuma K, Ma XX, Yuzawa H, Hiramatsu K. Insights on antibiotic resistance of Staphylococcus aureus from its whole genome: genomic island SCC. Drug Resist Updat 2003;6(1):41-52. doi: 10.1016/s1368-7646(03)00003-7. [Crossref]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec , encodes methicillin resistance in Staphylococcus aureus . Antimicrob Agents Chemother 2000;44(6):1549-55. doi: 10.1128/ aac.44.6.1549-1555.2000. [Crossref]
- Moon JS, Lee AR, Kang HM, Lee ES, Kim MN, Paik YH, et al. Phenotypic and genetic antibiogram of methicillin-resistant staphylococci isolated from bovine mastitis in Korea. J Dairy Sci 2007;90(3):1176-85. doi: 10.3168/jds.S0022- 0302(07)71604-1. [Crossref]
- Cookson BD, Robinson DA, Monk AB, Murchan S, Deplano A, de Ryck R, et al. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J Clin Microbiol 2007;45(6):1830-7. doi: 10.1128/ jcm.02402-06. [Crossref]
- Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus . Trends Microbiol 2001;9(10):486-93. doi: 10.1016/s0966- 842x(01)02175-8. [Crossref]
- Ito T, Katayama Y, Hiramatsu K. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob Agents Chemother 1999;43(6):1449-58.
- Ma XX, Ito T, Tiensasitorn C, Jamklang M, Chongtrakool P, Boyle-Vavra S, et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob Agents Chemother 2002;46(4):1147-52. doi: 10.1128/ aac.46.4.1147-1152.2002. [Crossref]
- Sadeghi Y, Salami SA, Kananizadeh P, Mozhgani SH, Pourmand MR. Real-time PCR followed by high-resolution melting analysis - a new robust approach to evaluate SCCmec typing of methicillin-resistant Staphylococcus aureus . Future Microbiol 2019;14:155-64. doi: 10.2217/fmb-2018-0193. [Crossref]
- Oliveira DC, Milheirico C, de Lencastre H. Redefining a structural variant of staphylococcal cassette chromosome mec , SCCmec type VI. Antimicrob Agents Chemother 2006;50(10):3457-9. doi: 10.1128/aac.00629-06. [Crossref]
- Zhang K, McClure JA, Elsayed S, Conly JM. Novel staphylococcal cassette chromosome mec type, tentatively designated type VIII, harboring class A mec and type 4 ccr gene complexes in a Canadian epidemic strain of methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 2009;53(2):531-40. doi: 10.1128/aac.01118-08. [Crossref]
- Ghaznavi-Rad E, Fard-Mousavi N, Shahsavari A, Japoni-Nejad A, Van Belkum A. Distribution of staphylococcal cassette chromosome mec types among methicillin-resistant coagulase negative staphylococci in central Iran. Iran J Microbiol 2018;10(1):7-13.
- Classification of staphylococcal cassette chromosome mec (SCCmec ): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 2009;53(12):4961-7. doi: 10.1128/aac.00579-09. [Crossref]
- Oliveira DC, Tomasz A, de Lencastre H. The evolution of pandemic clones of methicillin-resistant Staphylococcus aureus : identification of two ancestral genetic backgrounds and the associated mec elements. Microb Drug Resist 2001;7(4):349-61. doi: 10.1089/10766290152773365. [Crossref]
- Ito T, Katayama Y, Asada K, Mori N, Tsutsumimoto K, Tiensasitorn C, et al. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 2001;45(5):1323-36. doi: 10.1128/aac.45.5.1323-1336.2001. [Crossref]
- Lim TT, Chong FN, O’Brien FG, Grubb WB. Are all community methicillin-resistant Staphylococcus aureus related? a comparison of their mec regions. Pathology 2003;35(4):336- 43.
- Noto MJ, Archer GL. A subset of Staphylococcus aureus strains harboring staphylococcal cassette chromosome mec (SCCmec ) type IV is deficient in CcrAB-mediated SCCmec excision. Antimicrob Agents Chemother 2006;50(8):2782-8. doi: 10.1128/aac.00032-06. [Crossref]
- Goh SH, Byrne SK, Zhang JL, Chow AW. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol 1992;30(7):1642-5.
- Yang JA, Park DW, Sohn JW, Kim MJ. Novel PCR-restriction fragment length polymorphism analysis for rapid typing of staphylococcal cassette chromosome mec elements. J Clin Microbiol 2006;44(1):236-8. doi: 10.1128/jcm.44.1.236- 238.2006. [Crossref]
- Nasonova ES. [Pulsed field gel electrophoresis: theory, instruments and applications]. Tsitologiia 2008;50(11):927- 35.
- Ishino K, Tsuchizaki N, Ishikawa J, Hotta K. Usefulness of PCR-restriction fragment length polymorphism typing of the coagulase gene to discriminate arbekacin-resistant methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol 2007;45(2):607-9. doi: 10.1128/jcm.02099-06. [Crossref]
- Oliveira CF, Paim TG, Reiter KC, Rieger A, D’Azevedo PA. Evaluation of four different DNA extraction methods in coagulase-negative staphylococci clinical isolates. Rev Inst Med Trop Sao Paulo 2014;56(1):29-33. doi: 10.1590/s0036- 46652014000100004. [Crossref]
- Adaleti R, Nakipoglu Y, Karahan ZC, Tasdemir C, Kaya F. Comparison of polymerase chain reaction and conventional methods in detecting methicillin-resistant Staphylococcus aureus . J Infect Dev Ctries 2008;2(1):46-50. doi: 10.3855/ jidc.321. [Crossref]
- Amiri Z, Safari D, Mousavi G. SCCmec gene cassette chromosome complex molecular characterization and typing of methicillin-resistant Staphylococcus aureus strains isolated from clinical samples martyr Beheshti Hospital during the year 1388. Quarterly J Grace 2010;14:439-46. [Persian].
- Abdollahi A, Koohpayeh SA, Najafipoor S, Mansoori Y, Abdollahi S, Jaafari S. Evaluation of drug resistance and staphylococcal cassette chromosome (SCCmec ) types among methicillin-resistant Staphylococcus aureus (MRSA). Alborz University Medical Journal 2012;1(1):47-52. doi: 10.18869/ acadpub.aums.1.1.47. [Persian]. [Crossref]
- Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, Khoon LY, Aziz MN, Hamat RA, et al. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol 2010;48(3):867-72. doi: 10.1128/jcm.01112-09. [Crossref]
- Maleki A, Ghafourian S, Taherikalani M, Soroush S. Alarming and threatening signals from health centers about multi drug resistance Staphylococcus haemolyticus . Infect Disord Drug Targets 2019;19(2):118-27. doi: 10.2174/187152651866618 0911142806. [Crossref]
- Mohammadi S, Sekawi Z, Monjezi A, Maleki MH, Soroush S, Sadeghifard N, et al. Emergence of SCCmec type III with variable antimicrobial resistance profiles and spa types among methicillin-resistant Staphylococcus aureus isolated from healthcare- and community-acquired infections in the west of Iran. Int J Infect Dis 2014;25:152-8. doi: 10.1016/j. ijid.2014.02.018. [Crossref]
- Japoni-Nejad A, Rezazadeh M, Kazemian H, Fardmousavi N, van Belkum A, Ghaznavi-Rad E. Molecular characterization of the first community-acquired methicillin-resistant Staphylococcus aureus strains from Central Iran. Int J Infect Dis 2013;17(11):e949-54. doi: 10.1016/j.ijid.2013.03.023. [Crossref]
- Thong KL, Junnie J, Liew FY, Yusof MY, Hanifah YA. Antibiograms and molecular subtypes of methicillin-resistant Staphylococcus aureus in local teaching hospital, Malaysia. J Microbiol Biotechnol 2009;19(10):1265-70.
- Chongtrakool P, Ito T, Ma XX, Kondo Y, Trakulsomboon S, Tiensasitorn C, et al. Staphylococcal cassette chromosome mec (SCCmec ) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother 2006;50(3):1001-12. doi: 10.1128/aac.50.3.1001- 1012.2006. [Crossref]
- Katayama Y, Robinson DA, Enright MC, Chambers HF. Genetic background affects stability of mecA in Staphylococcus aureus. J Clin Microbiol 2005;43(5):2380-3. doi: 10.1128/ jcm.43.5.2380-2383.2005. [Crossref]
- Merlino J, Watson J, Rose B, Beard-Pegler M, Gottlieb T, Bradbury R, et al. Detection and expression of methicillin/ oxacillin resistance in multidrug-resistant and non-multidrug-resistant Staphylococcus aureus in Central Sydney, Australia. J Antimicrob Chemother 2002;49(5):793-801. doi: 10.1093/ jac/dkf021. [Crossref]
- Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 2003;9(8):978- 84. doi: 10.3201/eid0908.030089. [Crossref]
- Daum RS, Ito T, Hiramatsu K, Hussain F, Mongkolrattanothai K, Jamklang M, et al. A novel methicillin-resistance cassette in community-acquired methicillin-resistant Staphylococcus aureus isolates of diverse genetic backgrounds. J Infect Dis 2002;186(9):1344-7. doi: 10.1086/344326. [Crossref]
- Francois P, Renzi G, Pittet D, Bento M, Lew D, Harbarth S, et al. A novel multiplex real-time PCR assay for rapid typing of major staphylococcal cassette chromosome mec elements. J Clin Microbiol 2004;42(7):3309-12. doi: 10.1128/jcm.42.7.3309- 3312.2004. [Crossref]
- Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus . Antimicrob Agents Chemother 2002;46(7):2155-61. doi: 10.1128/aac.46.7.2155-2161.2002. [Crossref]