Avicenna Journal of Clinical Microbiology and Infection. 6(2):66-74.
doi: 10.34172/ajcmi.2019.13
Research Article
Isolation of Lactobacillus plantarum Strains With Robust Antagonistic Activity, Qualified Probiotic Properties, and Without Antibiotic-resistance From Traditional Sourdough
Mina Zangeneh 1  , Moj Khaleghi 1, *
, Moj Khaleghi 1, *  , Sadegh Khorrami 2
, Sadegh Khorrami 2 
Author information:
1Department of Biology, Faculty of Science, Shahid Bahonar University of Kerman, Kerman, Iran
2Department of Biotechnology, Faculty of Advanced Sciences and Technologies, University of Isfahan, Isfahan, Iran
*
Corresponding author: Moj Khaleghi, Tel: +983433257141-5, Fax: +983433257165, Email:
m.khaleghi@uk.ac.ir
Abstract
Background: The main purpose of the present study was to identify the qualified probiotic strains of traditional sourdough and to find stable Lactobacillus strains with enhanced antibacterial/antagonistic activity and without antibiotic resistance.
Methods: Sixteen strains were isolated from traditional sourdough and their susceptibility was evaluated toward a number of different antibiotics. In addition, the isolated strains were assessed with respect to their probiotic properties, antagonistic activity, and resistance to low pH, bile salt, different NaCl concentrations, and low storage temperatures.
Results: Based on the results, although none of the strains had considerable antibiotic resistance, they showed significant antagonistic activity against 6 different pathogens. Among the isolates, H3a and K3b were the most resistant to low pH and bile salt (0.3%) and demonstrated most hydrophobicity, auto-aggregation, and coaggregation characteristics. It was also observed that the strains could produce strong biofilm and were viable at 1%-10% NaCl concentrations, as well as at 4° C and -20° C. Finally, the sequencing results revealed that these 2 strains had the highest homology to Lactobacillus plantarum based on the 16S rRNA gene.
Conclusions: The present data confirmed the safety of Lactobacillus content of the traditional sourdough and the unique probiotic properties of these strains.
Keywords: Antagonistic activity, Antibiotic resistance, Lactobacillus plantarum, Probiotic, Sourdough
Copyright and License Information
© 2019 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Introduction
Researchers believe that food plays an increasingly important role in society with respect to disease prevention and human well-being. For this reason, the microbial aspects of food safety should seriously be taken into consideration (1,2). Due to their essential role in producing flavor and providing food preservation, lactic acid bacteria (LAB, e.g., lactobacilli) are highly important in fermented foods. It is found that the use of these bacteria as probiotic starters leads to the production of stable intestinal flora, competition with potentially harmful bacteria, and the prevention of certain diseases (3-5). In addition, these bacteria are considered as effective means for enhancing food safety and stability due to consumers’ awareness and concern regarding chemical additives (6). Further, LAB can act as an antagonist against some pathogenic bacteria by producing various antimicrobial compounds such as bacteriocins (7), organic acids, hydrogen peroxide, carbon dioxide, and diacetyl (8,9). Similarly, these diverse phylogenetic groups can be found in a wide variety of natural media, including fermented food products (e.g., yogurt, cheese, sauerkraut, coffee, sausages, vegetables, sourdough, and kimchi), as well as the normal flora of the mouth, gut and the vagina of the mammals (10-12)
To confer health benefits, probiotics need to survive in the gut and to attach and colonize the intestine. Therefore, they must survive in the acidic conditions of the stomach, as well as the acidic environment of some fermented foods and tolerate bile salt in the small intestine for colonization and metabolic activity (13). Furthermore, it is very important that they can maintain their viability during the production, processing, and storage of the fermented products (14). On the other hand, the risk of the transmission of antibiotic resistance genes to pathogenic and commensal bacteria in the gut should seriously be taken into consideration by using probiotics in the foods (15–17). Thus, the antibiotic resistance of different probiotic strains should be identified for safety reasons.
Accordingly, the isolation, identification, and characterization of native food born strains are regarded as the most important issues of national food industries given the importance of local foods and their striking role in public health. Although there are different bacterial species in sourdough, lactobacilli are the most abundant ones in this fermented food (10). Therefore, the present study aimed to assess the antibiotic resistance and antagonistic activity of native Lactobacillus strains with potential probiotic properties isolated from traditional sourdough of Kermanshah Province, West Iran. In addition, the study focused on evaluating the stability and growth rate of the isolated strains in sodium chloride (1%-10% w:v), as well as the effect of storage temperature of 4°C and -20°C on their growth rate.
Material and Methods
Materials
Mueller-Hilton Agar, Man Rogosa and Sharpe (MRS) media agar/broth, Anaerocult A-strip, HCl, n-hexadecane, and sodium chloride were purchased from Merck Company (Germany). In addition, antibiotic disks, as well as ethanol and acetic acid were obtained from the MAST Group (Merseyside, UK) and Sigma Aldrich (Germany), respectively. A genomic DNA extraction kit was procured from Bioneer Company (South Korea) as well. Finally, bile salt was purchased from Sigma Company (Oxgall, USA).
Samples and Initial Isolation of Lactobacilli Bacteria
Sixteen samples of traditional sourdough were collected in the Kermanshah province (West of Iran), Kermanshah during 2017, and were transferred to Shahid Bahonar University of Kerman, Kerman for further investigation. First, 10 g of each sample was added to 90 mL of sterile peptone water (1%) and homogenized for one minute by vortex mixer. Next, 10 dilutions of each sample were prepared in sterile peptone water and 100 µL of the dilutions were spread on the MRS agar media, followed by incubating for 48 hours at 37°C under anaerobic condition in a jar with Anaerocult A-strip. After initial tests, 16 gram-positive bacilli or coccobacilli and catalase-negative bacteria were chosen to study their most important probiotic criteria including antibiotic resistance, antagonistic activity, resistance to low pH, resistance to bile salt, biofilm formation, cell surface hydrophobicity, auto-aggregation and co-aggregation, and stability in storage conditions (18).
Antibiotic Resistance
In the present study, the antibiotic susceptibility of the isolated strains was tested toward different antibiotics including vancomycin (30 µg), penicillin (10 µg), erythromycin (15 µg), tetracycline (30 µg), gentamycin (10 µg), ceftriaxone (30 µg), imipenem (10 µg), oxacillin (1 µg), amoxicillin (10 µg), chloramphenicol (30 µg), and trimethoprim/sulfamethoxazole (1.25/23.75 µg). Briefly, the 0.5 McFarland suspensions of each 16 strains were prepared from their overnight cultures. Next, the suspensions were swabbed onto Mueller-Hinton agar plates and the antibiotic discs were embedded carefully. Then, the plates were incubated at the anaerobic condition at 37ºC for 24 hours. Subsequently, the diameters of the inhibition zones of each disc were measured and interpreted based on the guidelines of the Clinical Laboratory Standards Institute (CLSI) (19).
Antagonistic Activity
The agar well diffusion assay was used to test the antagonistic activity of the bacterial strains against 6 pathogen strains according to Khan and Kang (3). First, the cultures of Lactobacillus strains, which were grown overnight, were centrifuged at 9000 rpm for 15 minutes at 4°C using the Hettich, Mikro 200R centrifuge (UK) and the supernatant was filtered as well. Next, 107 CFU/mL of the target bacteria (i.e., Escherichia coli PTCC1330, Micrococcus luteus PTCC1110, Staphylococcus aureus PTCC1112, Bacillus subtilis PTCC1023, Enterococcus faecalis PTCC1237, and Listeria monocytogenes ATCC35152) were spread on the Mueller-Hilton Agar plates. The wells were then made on the plates using a clean and sterile borer, followed by placing 100 μL of the supernatant of the isolated Lactobacillus strains inside the wells and incubating the plates overnight at 37°C. Finally, the diameter of the zone of inhibition was measured and recorded in mm.
Resistance to Low pH
The resistance of the isolated strains to low pH was evaluated by a commonly employed method according to (20) with some modifications. First, the overnight cultures were centrifuged and the pellets were washed, re-suspended in peptone saline, and approximately 108 CFU/mL of them were inoculated into MRS broth adjusted to pH = 3 and 4 utilizing HCl (1 N). During this stage, MRS broth (pH = 6.5) was applied as a control culture medium. The cultures were next incubated at 37°C under anaerobic conditions for 3 hours. Then, 50 µL of these cultures were spread on MRS agar and incubated at 37°C under anaerobic conditions for 24 hours. Eventually, the number of live bacteria was counted as well.
Resistance to Bile Salt
To evaluate the resistance of the isolated strains to bile salt, the overnight cultures of the bacterial strains were centrifuged and their resultant pellets were washed and resuspended in peptone saline. Then, approximately 108 CFU/mL of the suspensions were inoculated into MRS broth enriched with bile salt (0.3%, w/v). MRS broth without the bile salt was employed as a control medium in this step. After 3 hours of incubation at 37°C under anaerobic conditions, 50 µL of the suspension was spread on MRS agar and incubated at 37°C under anaerobic conditions for a further 24 hours. The viability of the cells was finally determined using the colony count method.
Biofilm Formation
The biofilm formation was assessed using 96 well microplates according to the method previously described by our group (21) with some modifications. In the beginning, 250 µL of fresh MRS broth containing 108 CFU/mL of each of the isolated strains in the stationary phase was distributed in the wells and then the plates were incubated under the anaerobic condition at 30°C for 24 hours. After incubation, the medium was removed from each well, and the plates were washed twice with 250 μL of physiological saline solution to remove the free cells. Next, 250 µL of 96% ethanol (v/v) was added to each well to fix the formed biofilm. Then, the plates were dried and stained by adding 200 µL crystal violet (2% v/v) to each well and washing them twice after 30 minutes. Subsequently, the stained biofilms were dissolved by adding 200 µL acid acetic (33% v/v) and then the UV-Vis absorbance of each well at 490 nm was determined using a microplate reader (Bio-Rad Laboratories, Inc., USA). Three wells containing sterile MRS broth medium, as well as a Staphylococcus aureus strain were considered as negative controls and the positive control in each plate, respectively.
Cell Surface Hydrophobicity
Bacterial adhesion to hydrocarbons was conducted based on the method described by Todorov et al (22). The isolated strains were anaerobically grown in MRS broth at 37°C for 18 hours. Next, the culture media were centrifuged (at 9000 rpm at 4°C for 6 minutes), and the harvested cells were washed twice with quarter-strength Ringer’s solution. The cells were then resuspended in the same solution and their optical density (OD580 nm) was measured using a spectrophotometer (JASCO V-670 UV-VIS-NIR Spectrophotometer, Tokyo, Japan). Afterward, a mixture of 1.5 mL n-hexadecane was added to 1.5 mL cell suspension and vortexed for 2 minutes. Then, the mixture was allowed to separate its phases for 30 minutes at room temperature and the optical density (OD580 nm) of 1 mL of the aqueous phase was measured as well. In addition, the experiment was performed in triplicate, followed by determining the average optical density value. Finally, the percentage of hydrophobicity was calculated as follow:
%Hydrophobicity = ((A0 – A)/A0) ×100
where A0 and A are the absorbance values measured before and after n-hexadecane extraction, respectively.
Auto-aggregation and Co-aggregation Assay
To measure the auto-aggregation ability of the strains, the method reported by Khan and Kang was used with some modifications (3). To this end, 107 cells/mL of overnight cultures were centrifuged and then the pellets were washed 3 times with physiological saline solution, followed by resuspending them in 2 mL of fresh physiological saline solution. Next, the OD600 nm of cell suspension was monitored after 0, 0.5, 1, 2, 3, 4, and 5 hours of incubation at room temperature. Finally, the auto-aggregation ability was expressed as an auto-aggregation percentage using the following equation:
% Auto-aggregation = (1 – (At /Ainitial)) × 100
where At and Ainitial represent the absorbance at time t (i.e., 0.5, 1, 2, 3, 4, and 5 hours) and time 0, respectively.
The co-aggregation of the isolated strains with 6 pathogens was also determined similar to auto-aggregation. Briefly, the same volume of each Lactobacillus and pathogen strains were mixed and incubated at room temperature for 5 hours and the co-aggregation percentages were monitored after incubation according to the previously described method (3).
The co-aggregation was calculated using the following equation:
%Co-aggregation= ((OD Tot – OD S)/ OD Tot) ×100
OD Tot value refers to the initial OD immediately after the pairing of isolates and ODS denotes the OD of supernatant after 5 hours at room temperature.
Stability of Isolated Strains on Storage Conditions
In this study, 2 conditions of food storage were investigated, including different NaCl concentrations and low temperatures as follows.
-
The overnight cultures of the strains (approximately 108 CFU/mL) were inoculated in the MRS broth containing 0, 1, 2, 4, 6, 8, 10, and 12% (w/v) sodium chloride (NaCl). After 24 hours of anaerobic incubation, the salt the tolerance of the strains was determined by the colony count method (11)including tolerance to simulated gastric juice and bile salts, antimicrobial activity, antibiotic susceptibility and in vitro aggregation. Based on their morphological and biochemical characteristics, four potential probiotic isolates (CS2, CS3, CS4, and CS7.
-
The overnight cultures of each strain (approximately 108 CFU/mL) in MRS broth were stored at 4°C and -20ºC. One mL samples stored at 4°C and -20ºC were diluted and cultured on MRS agar. Eventually, the bacterial colonies were counted at the days of 1, 7, 14, 21, and 30 after 24 hours of incubation at 37ºC under anaerobic conditions (Figure 1).
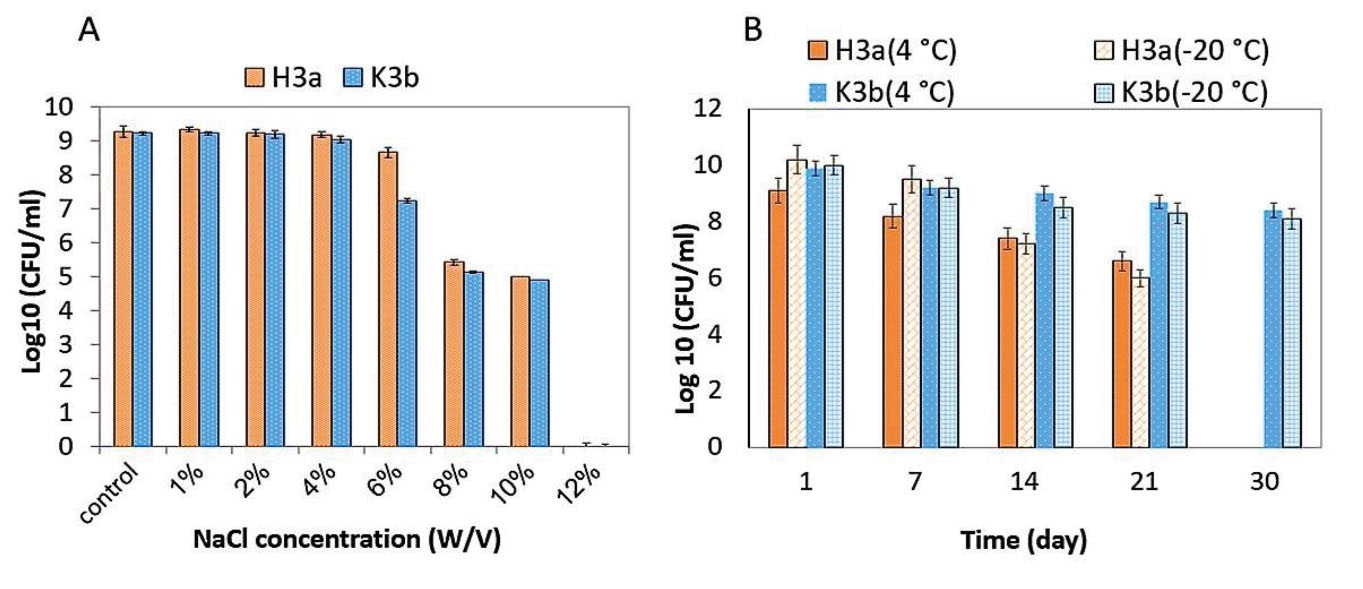
Figure 1.
Survival of H3a and K3b Strains in (A) Different NaCl Concentrations; (B) Low Storage Temperatures (4°C and -20ºC)
.
Survival of H3a and K3b Strains in (A) Different NaCl Concentrations; (B) Low Storage Temperatures (4°C and -20ºC)
Identification of the Isolates
Molecular identification of the selected strains as probiotic was confirmed by the sequencing of 16S rRNA. To extract bacterial genomic DNA, the genomic DNA extraction kit (Bioneer Co., Korea) was used based on the manufacturer’s instructions. Universal 16S rRNA polymerase chain reaction (PCR) primers set including U8F (5’-AGAGTTTGATCCTGGCTCAG-3’) as the forward primer and U1390R (5’-GACGGGCGGTGTGTACAA-3’) as the reverse primer were utilized to amplify 16S rRNA gene. The PCR was accomplished in a total volume of 50 µL including 50 ng of genomic DNA, 1.25 units of Taq DNA polymerase, 20 pmol of each primer, 19 PCR buffer and 200 µM of each dNTPs as the components. The PCR was performed as the following: for 35 cycles with primary denaturing for 3 minutes at 94°C, recurring denaturing for 30 seconds at 94°C, recombining in the double-stranded form for 30 seconds at 58°C, and extension for 2 minutes at 72°C with a final extension of 7 minutes at 72°C. Next, PCR products were electrophoresed on agarose gel (1%) and then the amplified 16S rRNA bands were purified by DNA extraction kit. In addition, DNA sequencing was directly conducted on both strands using a master mix provided by Bioneer Company (South Korea). The sequence data of 16S rRNA were then utilized for BLAST ((http://www.ncbi.nlm.nih.gov/BLAST/) analysis and the neighbor-joining phylogenetic tree was made using the Molecular Evolutionary Genetics Analysis Software, version 4.0 (23).
Statistical Analyses
All experiments were conducted in triplicate and the results were expressed as mean and standard deviation (SD). The values in the tolerance test were compared by one-way ANOVA and P values less than 0.05 were considered as a significant difference.
Results
Initial Isolation of Lactobacilli Strains
Initially, 16 strains were isolated from the 16 collected sourdough samples and were then studied with regard to their colony morphology, gram staining, and catalase production. Based on the results of this section, 16 strains were selected for further studies, including gram-positive bacilli/coccobacilli and catalase-negative. Figure 2 illustrates the purity and gram-positivity of one of the isolated Bacillus strains.
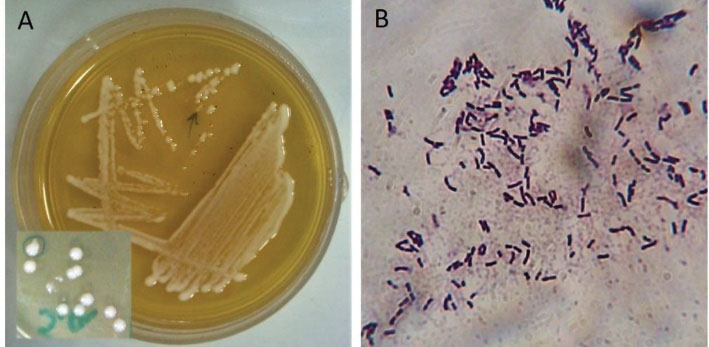
Figure 2.
(A)A Streak Culture and the Morphology of Colonies of One of the Isolated Strains (K3b); (B) Gram Staining of the K3b Strain. Note. The purity and Gram-positivity of the Bacillus strain are clear in this figure.
.
(A)A Streak Culture and the Morphology of Colonies of One of the Isolated Strains (K3b); (B) Gram Staining of the K3b Strain. Note. The purity and Gram-positivity of the Bacillus strain are clear in this figure.
Antibiotic Resistance Evaluation
The antibiotic resistance state of the isolated strains was evaluated regarding the 11 common antibiotics and based on the results, none of the isolated Lactobacillus strains possessed considerable antibiotic resistance toward the antibiotics. As shown in Table 1, the strains in the best way were resistance to only 4 out of 11 tested antibiotics. Vancomycin, oxacillin, and gentamicin were the most inactive antibiotics against the strains, respectively.
Table 1.
Antibiotic Resistance Pattern of the Isolated Strains
|
Strains
|
Susceptibility (S), Resistance (R), and Intermediate (I)
|
|
VAN
|
PEN
|
AMX
|
ERY
|
TET
|
GEN
|
CRO
|
SXT
|
IPM
|
CHL
|
OXA
|
| m3a |
R |
I |
S |
S |
S |
S |
S |
S |
S |
S |
I |
| H3a |
R |
S |
S |
S |
S |
S |
S |
S |
S |
S |
R |
| SH1a |
R |
I |
S |
S |
S |
I |
S |
R |
S |
S |
R |
| H3b |
R |
I |
S |
S |
S |
R |
S |
S |
I |
S |
R |
| SH2c |
R |
S |
S |
S |
S |
S |
R |
R |
S |
S |
R |
| D3 |
R |
S |
S |
S |
S |
S |
R |
S |
S |
S |
S |
| HE2a |
R |
S |
S |
S |
S |
S |
S |
S |
S |
S |
I |
| K10a |
R |
S |
S |
S |
S |
S |
S |
R |
S |
S |
S |
| A1d |
R |
R |
S |
S |
I |
S |
S |
S |
S |
S |
R |
| K3b |
R |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
| H1b |
R |
R |
S |
S |
S |
R |
S |
S |
S |
S |
R |
| ML4d |
R |
I |
S |
S |
S |
S |
R |
I |
S |
S |
R |
| K11d |
R |
I |
S |
S |
S |
R |
S |
S |
S |
S |
R |
| V3a |
R |
I |
S |
S |
S |
R |
S |
S |
S |
S |
R |
| K8d |
R |
R |
S |
S |
S |
S |
S |
S |
S |
S |
R |
| K11b |
R |
S |
S |
S |
S |
S |
S |
S |
S |
S |
S |
Note. Antibiotics: VAN, Vancomycin; PEN, Penicillin; AMX, Amoxicillin; ERY: Erythromycin; TET, Tetracycline; GEN, Gentamycin; CRO: Ceftriaxone; SXT, Trimethoprim/Sulfamethoxazole; IPM: Imipenem; CHL, Chloramphenicol; OXA, Oxacillin.
Antagonistic Activity of the Isolates
The antagonistic activity of the isolated strains was assessed against the 6 pathogenic bacteria. The evaluation revealed that almost all of the strains inhibit the growth of the pathogen. According to the results, 13 of the isolated strains (81%) led to the inhibition zone with ≥20 mm diameters. Further, the most antagonistic activity of the strains was observed against L. monocytogenes ATCC 35152, and S. aureus PTCC 25923. Finally, H3a and K3b strains had the highest inhibitory effect against all pathogenic bacteria (Figure 3).
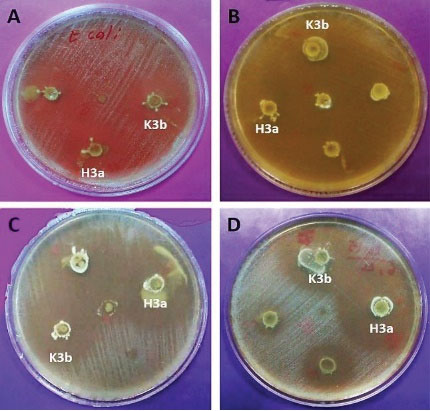
Figure 3.
Inhibitory Activity of K3b and H3a Strains Against (A) E. coli; (B) S. aureus; (C) E. faecalis; (D) L. monocytogenes.
.
Inhibitory Activity of K3b and H3a Strains Against (A) E. coli; (B) S. aureus; (C) E. faecalis; (D) L. monocytogenes.
Tolerance to Low pH and Bile Salt
Resistance to the low pH (3 & 4) of the isolated strains and bile salt (0.3%) was evaluated as well. Figure 4 displays the average colony count of the strains in these pH values and bile salt after 3 hours of exposure. The results indicate that 56.25% of the strains were resistant to pH = 3 although almost all strains were resistant to pH = 4. Moreover, 75% of the strains were resistant to bile salt. More precisely, V3a, K10a, H3a, H3b, and K3b strains could particularly survive and grow in both low pH and 0.3% bile salts.
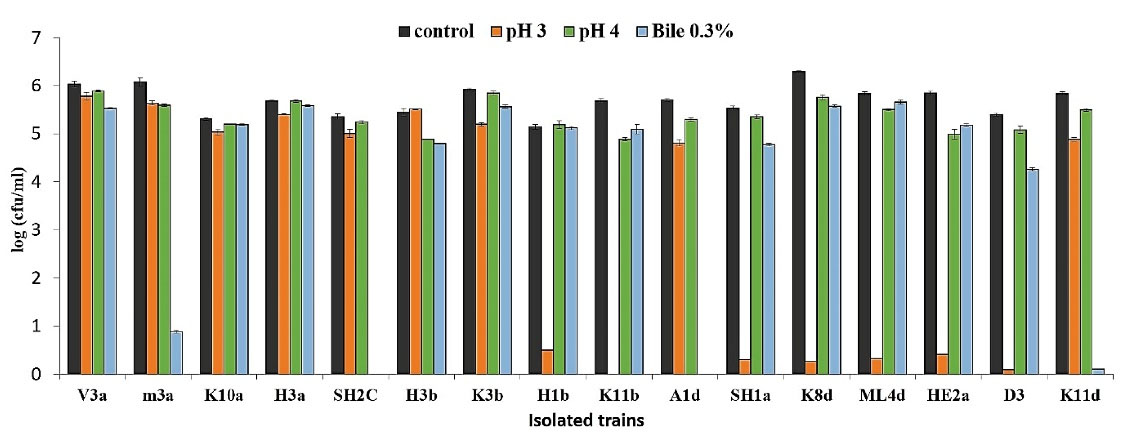
Figure 4.
The Average Colony Count of the Strains in the Low pH and Bile Salt After 3 Hours of Exposure
.
The Average Colony Count of the Strains in the Low pH and Bile Salt After 3 Hours of Exposure
Biofilm Formation
Biofilm formation was studied using the colorimetric method and based on the results, all 16 strains considerably produced biofilm compared to the control (S. aureus). However, H3a and K3b strains could form stronger biofilm compared to the control (Figure 5).
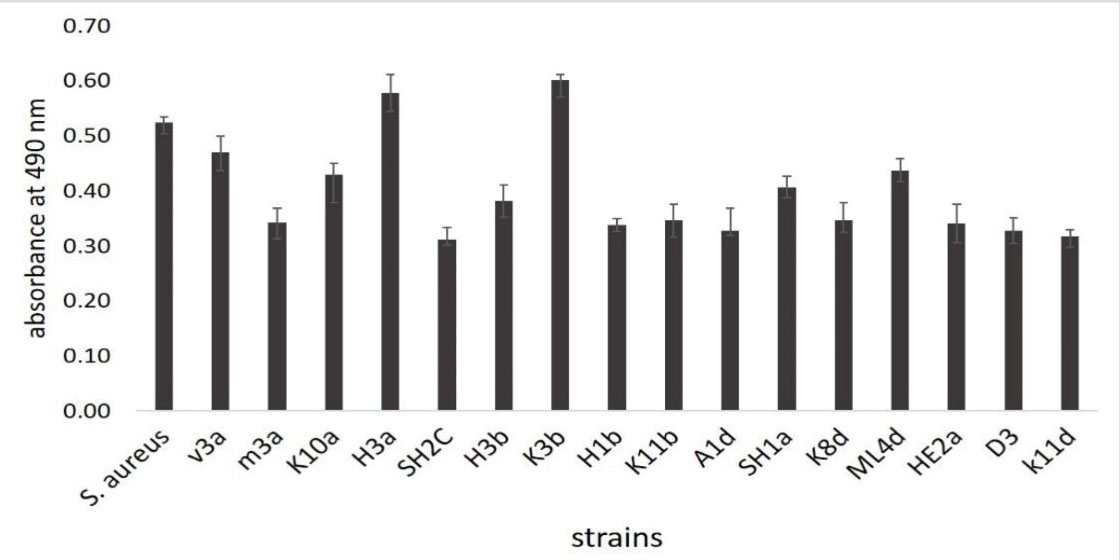
Figure 5.
Biofilm Formation of the Isolated Strains in Comparison to the Biofilm Formation by S. aureus.
.
Biofilm Formation of the Isolated Strains in Comparison to the Biofilm Formation by S. aureus.
Cell Surface Hydrophobicity
Considering the results of previous tests, H3a and K3b strains were selected for further study. Based on the surface hydrophobicity studies of both strains, H3a and K3b demonstrated 43% and 73% adhesion to n-hexadecane, respectively. N-hexadecane was used as a polar solvent and hydrophobic surface as well.
Aggregation Assay
The results of auto-aggregation and co-aggregation studies are depicted in Figure 6. According to slide A, the auto-aggregation of K3b (72%) during 5 hours of incubation at room temperature was significantly more than that of H3a (39%). In addition, the co-aggregate assessment results (Figure 6B) revealed that the ability of the co-aggregation of K3b with pathogens was more than that of H3a. Further, the K3b strain showed the highest rate of co-aggregation with S. aureus PTCC1112.
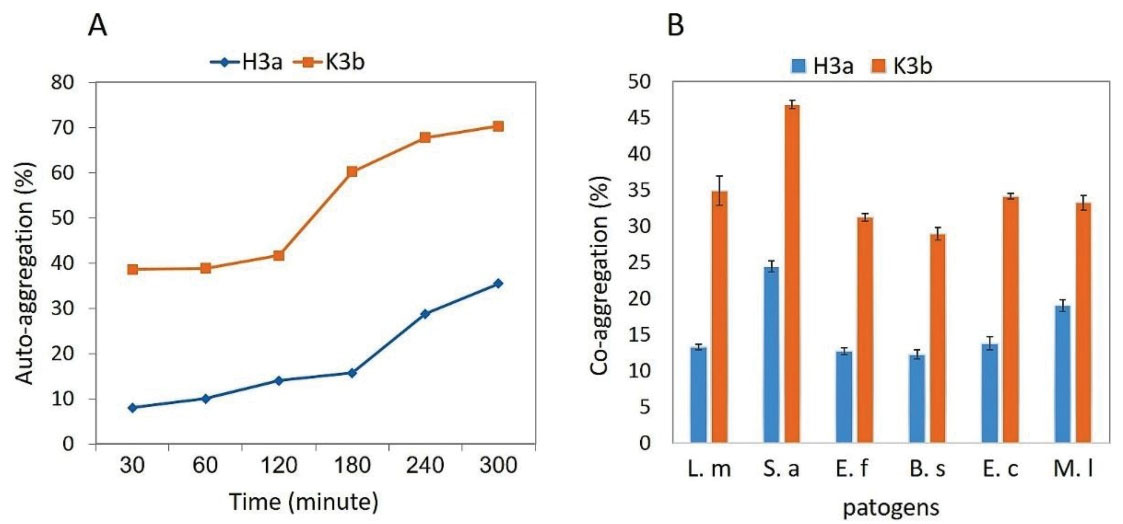
Figure 6.
(A) Auto-aggregation of H3a and K3b Strains in a 5-hour Period; (B) Co-aggregation Percent of Strains with Different Pathogens. Note. L. m: L. monocytogenes; S. a: S. aureus; E. f: E. faecalis; B. S: B. subtilis; E. c: E. coli; M. l: M. luteus.
.
(A) Auto-aggregation of H3a and K3b Strains in a 5-hour Period; (B) Co-aggregation Percent of Strains with Different Pathogens. Note. L. m: L. monocytogenes; S. a: S. aureus; E. f: E. faecalis; B. S: B. subtilis; E. c: E. coli; M. l: M. luteus.
Stability to NaCl and Low Temperature
The results of the growth and survival rate of the strains under the different concentrations of NaCl, as a harsh condition medium, showed that H3a and K3b strains were significantly resistant to 1%-10% NaCl concentrations. Contrarily, none of the strains could grow at 12% NaCl concentration (Figure 1A). Evaluating the survival of the strains at the storage temperatures of 4°C and -20°C represented that K3b strain was more resistant than H3a toward these temperatures (Figure 1B). The viability of this strain decreased to 3×108 CFU/mL and 1.3×108 CFU/mL at 4°C and -20°C, respectively, after one month of storage. H3a strain indicated partial survival at both temperatures for 3 weeks. However, its growth at 4°C and -20°C reduced to 3×106 CFU/mL and 3×105 CFU/mL, respectively.
Identification of the Isolates
The molecular identification of the 2 potent probiotic strains (i.e., H3a and K3b) was determined by using the information obtained from the sequencing of the 16S rRNA gene. Then, the phylogenetic tree was illustrated with MEGA4 software. Based on the results, H3a and K3b, which were identified as L. plantarum MZRF and L. plantarum KHMZ showed 99-100% similarity with L. plantarum WCFS1 and L. plantarum NBRC 15891, respectively (Figure 7).

Figure 7.
The Phylogenetic Tree Based on Partial 16S rDNA Sequences Representing the Relationship of H3a, uLb. plantarum MZRF (A) With K3b, uLb. plantarum KHMZ (B) Strains With Other Members of Lactobacillus spp. Note. The analysis was constructed using the neighbor-joining method in MEGA4.
.
The Phylogenetic Tree Based on Partial 16S rDNA Sequences Representing the Relationship of H3a, uLb. plantarum MZRF (A) With K3b, uLb. plantarum KHMZ (B) Strains With Other Members of Lactobacillus spp. Note. The analysis was constructed using the neighbor-joining method in MEGA4.
Discussion
Nowadays, the resistance of LAB to antibiotics has become a very important issue. It is assumed that some of the LAB may well transfer antibiotic resistance genes to other bacteria through horizontal gene transfer in the human gastrointestinal tract (24). The results of the present study also confirmed that the isolated strains were resistant to vancomycin, oxacillin, and gentamicin (Table 1), which is in line with the results of some studies regarding resistance to vancomycin and gentamicin (24,25). However, thankfully, the majority of the strains are still sensitive to the antibiotics, confirming the traditional sourdough as a healthy source. It is worth mentioning that the sensitivity of Lactobacillus strains toward erythromycin, tetracycline, ceftriaxone (26,27), and amoxicillin (28) was observed in earlier studies as well.
Although the antimicrobial activity of the probiotics against pathogenic bacteria was previously reported in the literature (9,29,30), the strains isolated in the present study demonstrated a special activity. The likelihood of this activity is high in producing various antimicrobial compounds, including sugar catabolites such as organic acids (i.e., lactic and acetic acids), fat and amino acid metabolites like fatty acids, phenyl lactic acid, and OH-phenyl lactic acid, as well as low-molecular-mass compounds such as hydrogen peroxide (H2O2), carbon dioxide (CO2), diacetyl (2,3-butanedione), and high-molecular-mass compounds such as bacteriocins (25).
The viability and survival during transit through the stomach and the upper part of the intestinal tract are considered as the most important indexes of a potent probiotic (31). According to evidence, the acid tolerance of LAB depends on the pH profile of H+-ATPase and the composition of the cytoplasmic membrane heavily relies on the type of bacteria, the type of growth media, and the conditions of their incubation (32). Furthermore, tolerance to bile salts is considered to be important for the colonization and metabolic activity of bacteria in the small intestine of the host. In addition, the bile resistance of some strains is due to the presence of bile salt hydrolase enzyme which hydrolyzes the conjugated bile in order to reduce its toxic effect (33). Our study showed that H3a and K3b strains were resistant to the low pH values and the employed bile salt condition so that their survival was more than 90% in the environment which was similar to gastric juice. Previous studies have similarly indicated that different isolated Lactobacillus strains were resistant to low pH and bile salt (13,20,34).
Likewise, adhesion is regarded as a complex process that involves non-specific (hydrophobicity) and specific ligand-receptor mechanisms (35). Further, biofilm is known as a microbial adherent structure and is associated with some diseases. In contrast, a beneficial biofilm of Lactobacilli probably plays a protective role in the gastrointestinal tract (6). Recent studies have reported that biofilm formation and its properties in Lactobacillus strains depend on the strain, the composition of the growth medium, and growth conditions (8,36). In the present study, it was observed that various Lactobacillus strains formed biofilms with different densities, which is in agreement with the results of previous research (37). However, H3a and K3b strains could form stronger biofilms.
Bacteria can adhere to either similar bacteria (auto-aggregation) or genetically different strains (co-aggregation) through the aggregation process. This process may play a significant role in eliminating the pathogens from the human gut (22,25). Collado et al found that bacterial surface characteristics and their adhesion ability to hydrocarbons correlate with their aggregation abilities (35). On the other hand, earlier studies revealed that cell surface hydrophobicity depends on the presence of (glycol) proteinaceous material at the bacterial surface (26,35,38). Our results demonstrated that K3b not only shows the highest adhesion to hydrocarbons but also higher aggregation ability (auto-aggregation and co-aggregation) compared to the other tested strains.
Storage conditions such as salt concentrations and low temperature in the food industry are important for food quality maintenance and the survivability of probiotics. In some countries, different concentrations of NaCl (1-10% w/v) are used as a preservative in fermented food (e.g., cheese, pickles, and sourdough). However, probiotics should be durable in high NaCl concentrations in food due to the inhibitory effect of NaCl on some bacteria (11,14)including tolerance to simulated gastric juice and bile salts, antimicrobial activity, antibiotic susceptibility and in vitro aggregation. Based on their morphological and biochemical characteristics, four potential probiotic isolates (CS2, CS3, CS4, and CS7. In the current study, H3a and K3b strains could survive at 1-10% NaCl concentrations. It was also detected that the K3b strain was more viable than H3a in low storage temperatures. In a similar study, Wang et al. concluded that 3 Lactobacillus strains considerably survived after 21 days of storage at 4ºC (18).
Conclusions
The results of the present research showed that the native Lactobacillus strains isolated from traditional sourdough have a good situation regarding their antibiotic resistance. It implies that they not only are highly sensitive to a great number of antibiotics but have also an incredible antagonistic activity. Furthermore, they have a potent probiotic potential, can survive at 10% NaCl and low temperatures and can form a strong biofilm. Moreover, the strains are capable of aggregating with pathogen strains. Finally, these strains are proper candidates and, undoubtedly, can be used at industrial levels.
Ethical Approval
This article contains no studies with human participants or animals performed by any of the authors.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Funding
The authors declare that they received no funding for the present research.
References
- Havelaar AH, Brul S, de Jong A, de Jonge R, Zwietering MH, Ter Kuile BH. Future challenges to microbial food safety. Int J Food Microbiol 2010; 139 Suppl 1:S79-94. doi: 10.1016/j.ijfoodmicro.2009.10.015 [Crossref] [ Google Scholar]
- Mor-Mur M, Yuste J. Emerging bacterial pathogens in meat and poultry: an overview. Food Bioproc Tech 2010; 3(1):24-35. doi: 10.1007/s11947-009-0189-8 [Crossref] [ Google Scholar]
- Khan I, Kang SC. Probiotic potential of nutritionally improved Lactobacillus plantarum DGK-17 isolated from Kimchi – A traditional Korean fermented food. Food Control 2016; 60:88-94. doi: 10.1016/j.foodcont.2015.07.010 [Crossref] [ Google Scholar]
- Riaz Rajoka MS, Shi J, Zhu J, Shao D, Huang Q, Yang H. Capacity of lactic acid bacteria in immunity enhancement and cancer prevention. Appl Microbiol Biotechnol 2017; 101(1):35-45. doi: 10.1007/s00253-016-8005-7 [Crossref] [ Google Scholar]
- Riaz Rajoka MS, Mehwish HM, Siddiq M, Haobin Z, Zhu J, Yan L. Identification, characterization, and probiotic potential of Lactobacillus rhamnosus isolated from human milk. LWT 2017; 84:271-80. doi: 10.1016/j.lwt.2017.05.055 [Crossref] [ Google Scholar]
- Castellano P, Belfiore C, Fadda S, Vignolo G. A review of bacteriocinogenic lactic acid bacteria used as bioprotective cultures in fresh meat produced in Argentina. Meat Sci 2008; 79(3):483-99. doi: 10.1016/j.meatsci.2007.10.009 [Crossref] [ Google Scholar]
- Vinderola G, Ouwehand A, Salminen S, von Wright A. Lactic Acid Bacteria: Microbiological and Functional Aspects. 5th ed. Boca Raton: CRC Press; 2019. 10.1201/9780429057465
- Aoudia N, Rieu A, Briandet R, Deschamps J, Chluba J, Jego G. Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: Effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol 2016; 53:51-9. doi: 10.1016/j.fm.2015.04.009 [Crossref] [ Google Scholar]
- Soltan Dallal MM, Davoodabadi A, Abdi M, Hajiabdolbaghi M, Sharifi Yazdi MK, Douraghi M. Inhibitory effect of Lactobacillus plantarum and Lb fermentum isolated from the faeces of healthy infants against nonfermentative bacteria causing nosocomial infections. New Microbes New Infect 2017; 15:9-13. doi: 10.1016/j.nmni.2016.09.003 [Crossref] [ Google Scholar]
- Corsetti A, Settanni L. Lactobacilli in sourdough fermentation. Food Res Int 2007; 40(5):539-58. doi: 10.1016/j.foodres.2006.11.001 [Crossref] [ Google Scholar]
- Jena PK, Trivedi D, Thakore K, Chaudhary H, Giri SS, Seshadri S. Isolation and characterization of probiotic properties of lactobacilli isolated from rat fecal microbiota. Microbiol Immunol 2013; 57(6):407-16. doi: 10.1111/1348-0421.12054 [Crossref] [ Google Scholar]
- Son SH, Jeon HL, Jeon EB, Lee NK, Park YS, Kang DK. Potential probiotic Lactobacillus plantarum Ln4 from kimchi: Evaluation of beta-galactosidase and antioxidant activities. LWT 2017; 85:181-6. doi: 10.1016/j.lwt.2017.07.018 [Crossref] [ Google Scholar]
- Muñoz-Quezada S, Chenoll E, Vieites JM, Genovés S, Maldonado J, Bermúdez-Brito M. Isolation, identification and characterisation of three novel probiotic strains (Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036) from the faeces of exclusively breast-fed infants. Br J Nutr 2013; 109 Suppl 2:S51-62. doi: 10.1017/s0007114512005211 [Crossref] [ Google Scholar]
- Gandhi A, Shah NP. Effect of salt on cell viability and membrane integrity of Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium longum as observed by flow cytometry. Food Microbiol 2015; 49:197-202. doi: 10.1016/j.fm.2015.02.003 [Crossref] [ Google Scholar]
- Adimpong DB, Nielsen DS, Sørensen KI, Derkx PM, Jespersen L. Genotypic characterization and safety assessment of lactic acid bacteria from indigenous African fermented food products. BMC Microbiol 2012; 12:75. doi: 10.1186/1471-2180-12-75 [Crossref] [ Google Scholar]
- Khorrami S, Zarrabi A, Khaleghi M, Danaei M, Mozafari MR. Selective cytotoxicity of green synthesized silver nanoparticles against the MCF-7 tumor cell line and their enhanced antioxidant and antimicrobial properties. Int J Nanomedicine 2018; 13:8013-24. doi: 10.2147/ijn.s189295 [Crossref] [ Google Scholar]
- Khorrami S, Jafari Najafabadi F, Zarepour A, Zarrabi A. Is Astragalus gossypinus Honey a Natural Antibacterial and Cytotoxic Agent? An Investigation on A gossypinus Honey Biological Activity and Its Green Synthesized Silver Nanoparticles. Bionanoscience 2019; 9(3):603-10. doi: 10.1007/s12668-019-00646-8 [Crossref] [ Google Scholar]
- Wang CY, Lin PR, Ng CC, Shyu YT. Probiotic properties of Lactobacillus strains isolated from the feces of breast-fed infants and Taiwanese pickled cabbage. Anaerobe 2010; 16(6):578-85. doi: 10.1016/j.anaerobe.2010.10.003 [Crossref] [ Google Scholar]
- Cockerill FR. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement. Clinical and Laboratory Standards Institute (CLSI); 2011.
- Huang Y, Wang X, Wang J, Wu F, Sui Y, Yang L. Lactobacillus plantarum strains as potential probiotic cultures with cholesterol-lowering activity. J Dairy Sci 2013; 96(5):2746-53. doi: 10.3168/jds.2012-6123 [Crossref] [ Google Scholar]
- Khaleghi M, Khorrami S, Ravan H. Identification of Bacillus thuringiensis bacterial strain isolated from the mine soil as a robust agent in the biosynthesis of silver nanoparticles with strong antibacterial and anti-biofilm activities. Biocatal Agric Biotechnol 2019; 18:101047. doi: 10.1016/j.bcab.2019.101047 [Crossref] [ Google Scholar]
- Todorov SD, Botes M, Guigas C, Schillinger U, Wiid I, Wachsman MB. Boza, a natural source of probiotic lactic acid bacteria. J Appl Microbiol 2008; 104(2):465-77. doi: 10.1111/j.1365-2672.2007.03558.x [Crossref] [ Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 40. Mol Biol Evol 2007; 24(8):1596-9. doi: 10.1093/molbev/msm092 [Crossref] [ Google Scholar]
- Murua A, Todorov SD, Vieira AD, Martinez RC, Cencič A, Franco BD. Isolation and identification of bacteriocinogenic strain of Lactobacillus plantarum with potential beneficial properties from donkey milk. J Appl Microbiol 2013; 114(6):1793-809. doi: 10.1111/jam.12190 [Crossref] [ Google Scholar]
- Bao Y, Zhang Y, Zhang Y, Liu Y, Wang S, Dong X. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010; 21(5):695-701. doi: 10.1016/j.foodcont.2009.10.010 [Crossref] [ Google Scholar]
- Zielińska D, Rzepkowska A, Radawska A, Zieliński K. In vitro screening of selected probiotic properties of Lactobacillus strains isolated from traditional fermented cabbage and cucumber. Curr Microbiol 2015; 70(2):183-94. doi: 10.1007/s00284-014-0699-0 [Crossref] [ Google Scholar]
- Naghizadeh Raeisi S, Ghoddusi HB, Juncker Boll E, Farahmand N, Stuer-Lauridsen B, Johansen E. Antimicrobial susceptibility of bifidobacteria from probiotic milk products and determination of the genetic basis of tetracycline resistance in Enterococcus species after in vitro conjugation with Bifidobacterium animalis subsp lactis. Food Control 2018; 94:205-11. doi: 10.1016/j.foodcont.2018.07.016 [Crossref] [ Google Scholar]
- Hoque MZ, Akter F, Hossain KM, Rahman MSM, Billah M, Islam KMD. Isolation, identification and analysis of probiotic properties of Lactobacillus spp from selective regional yoghurts. World J Dairy Food Sci 2010; 5(1):39-46. [ Google Scholar]
- Tulumoğlu Ş, Kaya Hİ, Şimşek Ö. Probiotic characteristics of Lactobacillus fermentum strains isolated from tulum cheese. Anaerobe 2014; 30:120-5. doi: 10.1016/j.anaerobe.2014.09.015 [Crossref] [ Google Scholar]
- Negi YK, Pandey C, Saxena N, Sharma S, Garg FC, Garg SK. Isolation of antibacterial protein from Lactobacillus spp and preparation of probiotic curd. J Food Sci Technol 2018; 55(6):2011-20. doi: 10.1007/s13197-018-3115-0 [Crossref] [ Google Scholar]
- Erkkilä S, Petäjä E. Screening of commercial meat starter cultures at low pH and in the presence of bile salts for potential probiotic use. Meat Sci 2000; 55(3):297-300. doi: 10.1016/s0309-1740(99)00156-4 [Crossref] [ Google Scholar]
- Rushdy AA, Gomaa EZ. Antimicrobial compounds produced by probiotic Lactobacillus brevis isolated from dairy products. Ann Microbiol 2013; 63(1):81-90. doi: 10.1007/s13213-012-0447-2 [Crossref] [ Google Scholar]
- Rani RP, Anandharaj M, Ravindran AD. Characterization of bile salt hydrolase from Lactobacillus gasseri FR4 and demonstration of its substrate specificity and inhibitory mechanism using molecular docking analysis. Front Microbiol 2017; 8:1004. doi: 10.3389/fmicb.2017.01004 [Crossref] [ Google Scholar]
- Wang L, Zhang H, Rehman MU, Mehmood K, Jiang X, Iqbal M. Antibacterial activity of Lactobacillus plantarum isolated from Tibetan yaks. Microb Pathog 2018; 115:293-8. doi: 10.1016/j.micpath.2017.12.077 [Crossref] [ Google Scholar]
- Collado MC, Meriluoto J, Salminen S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur Food Res Technol 2008; 226(5):1065-73. doi: 10.1007/s00217-007-0632-x [Crossref] [ Google Scholar]
- Fernández Ramírez MD, Smid EJ, Abee T, Nierop Groot MN. Characterisation of biofilms formed by Lactobacillus plantarum WCFS1 and food spoilage isolates. Int J Food Microbiol 2015; 207:23-9. doi: 10.1016/j.ijfoodmicro.2015.04.030 [Crossref] [ Google Scholar]
- Jones SE, Versalovic J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol 2009; 9:35. doi: 10.1186/1471-2180-9-35 [Crossref] [ Google Scholar]
- Bevilacqua A, Petruzzi L, Speranza B, Campaniello D, Sinigaglia M, Corbo MR. Changes of the cell surface hydrophobicity of Lactobacillus acidophilus La-5 in response to pH, temperature and inulin. Int J Food Sci Technol 2018; 53(5):1262-8. doi: 10.1111/ijfs.13706 [Crossref] [ Google Scholar]