Avicenna Journal of Clinical Microbiology and Infection. 10(4):162-165.
doi: 10.34172/ajcmi.3503
Original Article
Determining the Molecular Identity of Two Virulence Genes mec-A and lukS/F-PV of Staphylococcus aureus Isolated From the Lung Secretions of Patients Admitted to Intensive Care Units in Kerman, Iran
Mitra Zare 1  , Iman Pouladi 2
, Iman Pouladi 2  , Babak Kheirkhah 3, *
, Babak Kheirkhah 3, * 
Author information:
1Department of Microbiology, Faculty of Science, Kerman Branch, Islamic Azad University, Kerman, Iran
2Department of Microbiology and Immunology, Faculty of Veterinary Medicine, University of Tehran, Iran
3Department of Microbiology, Faculty of Veterinary Medicine, Baft Branch, Islamic Azad University, Baft, Iran
Abstract
Background: Staphylococcus aureus is one of the most common hospital- and community-acquired pathogens. This bacterium has different virulence factors, and today’s reports show that the prevalence of methicillin resistance in S. aureus is increasing in different regions of the world.
Methods: This cross-sectional descriptive study was performed on 60 hospital samples. First, biochemical tests were conducted on the samples to separate and confirm the genus of S. aureus. After the enrichment and isolation of bacteria and extraction of the DNA of mec-A and lukS/F-PV genes, they were evaluated by multiplex polymerase chain reactions (PCR).
Results: In this study, 20 isolates of S. aureus were obtained from a total of 60 samples. In these 20 S. aureus isolates, the frequency of the lukS/F-PV gene was reported in 12 isolates, and the frequency of the mecA gene was reported in 8 isolates, of which 4 isolates had both genes.
Conclusion: According to the results, the identification of genes related to the severity of the disease and antibiotic resistance can play an effective role in the identification of the antibiotic-resistant population and subsequent planning to deal with antibiotic resistance. In addition, multiplex PCR, as a low-cost and specific method, was used to identify infectious agents of pathogens related to their virulence.
Keywords: Staphylococcus aureus, Respiratory Tract infections, PCR, Virulence Factors, lukS/F-PV, mecA
Copyright and License Information
© 2023 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Zare M, Pouladi I, Kheirkhah B. Determining the molecular identity of two virulence genes mec-A and lukS/F-PV of Staphylococcus aureus isolated from the lung secretions of patients admitted to intensive care units in Kerman, Iran. Avicenna J Clin Microbiol Infect. 2023; 10(4):162-165. doi:10.34172/ajcmi.3503
Introduction
The skin, which is the first defense barrier of the body (1), is also the main site of staphylococci. Pathogenic staphylococci are quickly destroyed by skin secretions and other antimicrobial factors, but resistant staphylococci remain, which can cause severe infections depending on the number of bacteria. For example, in experimental conditions, many bacteria (i.e., more than 1 million) are required (2). Staphylococcus aureus is a nosocomial and community-acquired infection that harbors a variety of pathogens and provides host invasion requirements. Through the release of various toxins, superantigenic enterotoxin SEA and SEB are most likely to cause pathogenicity (3,4).
The amount of contamination will be greatly reduced if the skin has a lesion or a crack. In any case, deep infections of the body originate from the skin, and a few of these infections originate from the respiratory and administrative genital tracts (2). The infection is limited after the passage of bacteria from the lung into the tissue due to the secretion of coagulase and the formation of fibrin. Later, polymorphonucleotide leukocytes quickly come to the site of infection and swallow them, and as a result, the infection quickly becomes necrotic, and the bacteria and dead leukocytes gradually produce thick, cream-colored pus (5). Around the infection, a fibroblastic wall is created by the activity of fibroblasts. If the surrounding tissue cannot limit the infection, staphylococci may enter the blood vessels and cause septicemia or small boils in the lungs, kidneys, heart, bones, and under the skin (6). In humans, the main locations of S. aureus are the nasal cavities and the anterior part of the nostrils, where they are permanently present. In adults, this presence is about 50 %, while it is higher in children (6). It has been shown that 5‒30% people are infected with staphylococcus on their hands and faces (7). In addition to the nasal cavity, throat, and intestines, it is also found that germs enter the air and dust from these sources; and by contaminating clothes and working tools at the workplace, they cause infections in the lungs. Although the two most common sources of staphylococci are the nose and hands, it should be kept in mind that most pets can harbor S. aureus (5). The purpose of our research was to recognize the molecular identity of S. aureus virulence genes isolated from patients hospitalized in the intensive care units (7). The isolation of bacterial virulence genes by the polymerase chain reaction (PCR) method helps researchers and doctors better understand the pathogenesis of the disease and find ways to deal with this infection. The importance of S. aureus in strains, the creation of nosocomial infections, and the role of virulence genes of this bacterium were motivations to use the PCR method in this research (8). Therefore, this study aimed to investigate the molecular identity of the virulence genes of S. aureus isolated from the lung secretions of patients hospitalized in the special care departments of hospitals of Kerman.
Materials and Methods
Collection of Clinical Samples and Identification of Bacteria
In our cross-sectional descriptive study, 60 samples of lung secretions using swabs were taken from patients hospitalized in the special care department of Kerman hospitals. The swabs of the samples were placed into Stuart’s transport media and immediately transferred to the Microbiology Laboratory of Islamic Azad University, the Kerman branch. Bacterial identification was performed using standard microbiological methods, such as colony morphology, gram stain, catalase testing, coagulase assay, and Mannitol salt agar, and was confirmed by molecular methods.
Extraction DNA
DNA extraction was performed using the National Center for Genetic Resources kit according to the instructions provided by the Gram-positive bacteria DNA extraction kit.
Polymerase Chain Reaction Method to Identify 16S rRNA, mecA, and LukS /F-PV Genes
After extracting the genome of the samples, the PCR was performed to identify 16S rRNA, mec -A, and LukS /F-PV genes using specific primers (Table 1) in a final volume of 25 μL according to the protocol of one step of initial denaturation at 95 °C for 5 minutes, 40 cycles of denaturation at 95 °C for 40 seconds, annealing at 58 °C for 1 minute, and elongation at 72 °C for 2 minutes, and a final elongation step at 72 °C for 7 minutes.
Table 1.
Nucleotide Sequences and Primers Used in This Study
|
Gene
|
Primer
|
Primer Sequence
|
Product Size
|
Reference
|
|
16S rRNA
|
staph756F |
5ꞌ-AACTCTGTTATTAGGGAAGAACA-3ꞌ |
756 |
(9) |
| Staph750R |
5ꞌ-CCACCTTCCTCCGGTTTGTCACC-3ꞌ |
|
LukS /F-PV
|
Luk-PV-1 |
5ꞌ-ATCATTAGGTAAAATGTCTGGACATGATCCA-3ꞌ |
433 |
(10) |
| Luk-PV-2 |
5ꞌ- GCATCAATSGTATTGGATAGCAAAGC -3ꞌ |
|
mecA
|
mecA1 |
5ꞌ- GTAGAAATGACTGAACGTCCGATAA -3ꞌ |
310 |
(11) |
| mecA2 |
5ꞌ-CCAATTCCACATTGTTTCGGTCTAA-3ꞌ |
Then, the PCR in the final volume of 25 μL contained 12.5 μL of Master Mix, 1 μL of F primer, 1 μL of R primer, and 2 μL of bacterial DNA, including Primer-Fwd (100 Pm) – 1.5 μL (each primer 0.25), Primer-ReV (100 Pm) – 1.5 μL (each Primer 0.25), Tag DNA polymerase – 0.1, DNA template – 2.5 μL, total volume: 26.5 μL, and 8.5 μL of distilled water according to the protocol. It also contained an initial denaturation step at a temperature of 95 °C for 5 minutes, 40 cycles of denaturation at 95°C for 40 seconds, annealing at 58 °C for 45 seconds, and elongation at 72 °C for 1 minute, and a final elongation step at 72 °C for 10 minutes to identify mecA and LukS /F-PV genes (Table 1).
Results
The present study was conducted on 60 clinical samples of patients hospitalized in the special care department of Kerman hospitals. Bacteria were identified according to standard microbiological methods (morphology, gram staining, catalase test, coagulase test, and mannitol salt fermentation), and the identity of 20 S. aureus isolates was confirmed and then compared to DNA extraction and the PCR test using specific primers. It was performed for the mecA, 16S rRNA,and LukS /F-PV genes. After identifying 20 isolates of S. aureus according to standard microbiological methods, the 16S rRNA gene was positive as a reference gene for the identification of S. aureus species in 20 samples (Figure 1).
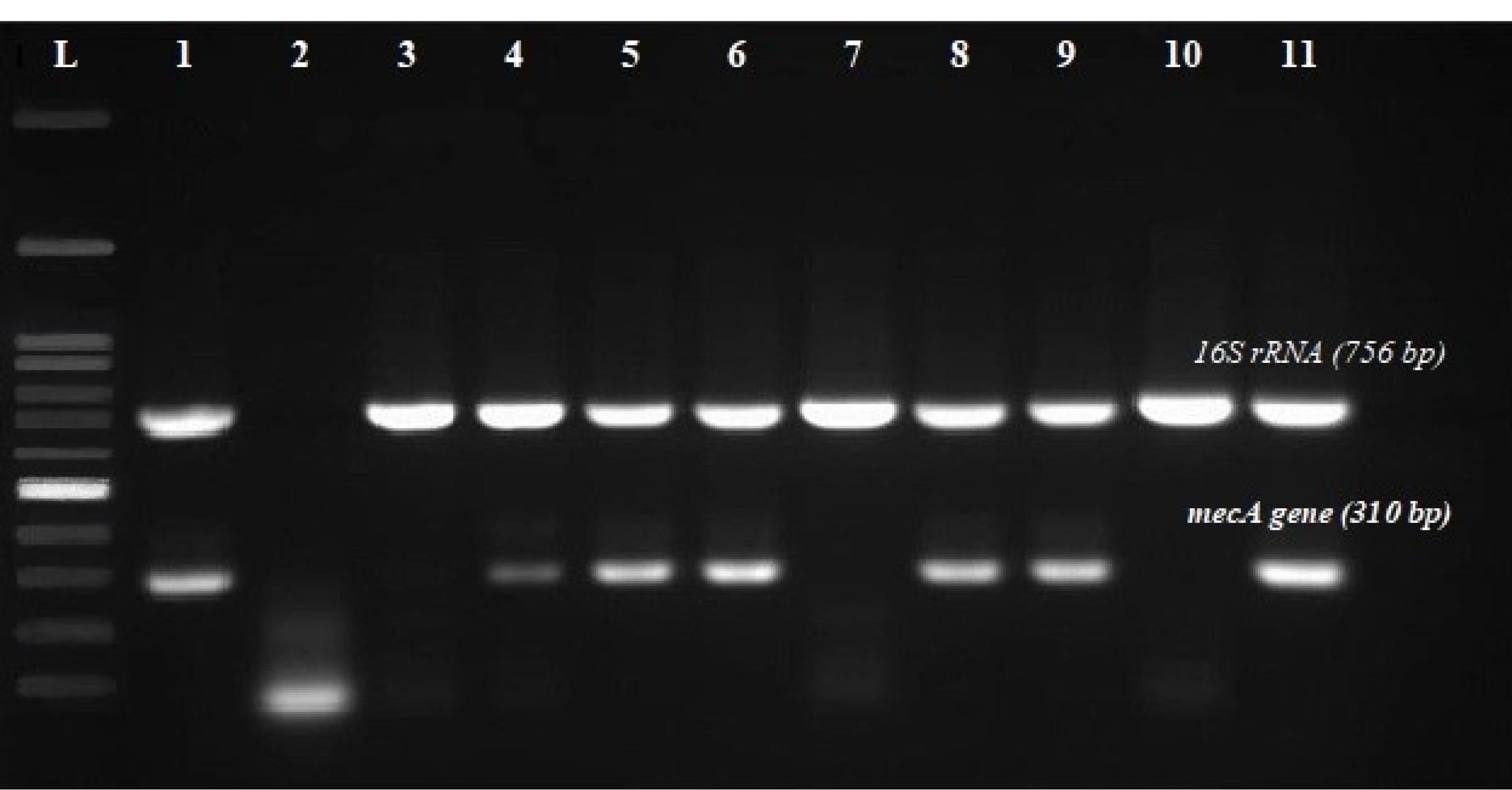
Figure 1.
Multiplex PCR Results. From the left, row (L): DNA Ladder 100 bp, Row (1): Positive control, Row (1): Negative control, Rows 3, 4, and 5 to 11: Strains carrying the gene are 16S rRNA (756 bp), Rows 4, 5, 6, 8, 9, and 11: Strains carrying the mecA gene (310 bp)
.
Multiplex PCR Results. From the left, row (L): DNA Ladder 100 bp, Row (1): Positive control, Row (1): Negative control, Rows 3, 4, and 5 to 11: Strains carrying the gene are 16S rRNA (756 bp), Rows 4, 5, 6, 8, 9, and 11: Strains carrying the mecA gene (310 bp)
In these 20 S. aureus isolates, the frequencies of the LukS /F-PV and mecA genes were reported in 12 and 8 isolates, respectively, of which 4 isolates had both genes (Table 2).
Table 2.
Frequencies and Percentages of Infection With Staphylococcus aureus
|
|
S. aureus
|
LUKS/F-PV
|
macA
|
|
Frequency
|
S. aureus
(%)
|
Frequency
|
Gene LukS/F-PV (%)
|
Frequency
|
macA
(%) Gene
|
| Positive |
20 |
33.33 |
12 |
19 |
8 |
13 |
| Negative |
40 |
66.67 |
48 |
81 |
52 |
87 |
| Total |
60 |
100 |
60 |
100 |
60 |
100 |
Discussion
Different bacterial species, especially the important species of S. aureus, Acinetobacter baumannii, and Pseudomonas aeruginosa, are common causes of hospital infections, especially in intensive care units, due to the presence of virulence genes. Considering that S. aureus causes various diseases in humans, the analysis of the pathogenic factors of this bacterium is of great importance (12-14). Overall, 20 isolates of S. aureus were obtained based on the results of this study. The frequency of LukS /F-PV and mecA genes was 24% and 36%, respectively. By examining the recent studies and comparing their results with those of the present study, there was no significant difference, and no incompatibility of the phenotypic and genotypic results was found in the studies of other researchers. Based on the results of the first comprehensive study by Najafi Olya et al, investigating the clonal diversity and genomic characteristics of positive S. aureus (PVL) in Iran, it was determined that PVL-positive MRSA strains are highly prevalent in Iran. The results of this study, compared to those of the present study, on the prevalence of methicillin-resistant S. aureus and the mec -A gene, lukS /FPV, may contribute to the understanding of molecular epidemiology and evolution (15). In a study by Trevizani Rocchetti et al, out of 85 samples (23.0%) of blood cultures, S. aureus and CoNS were detected in 286 samples (77.0%). A total of 85 (50.6%) S. aureus samples carried the mecA gene, and among 286 CoNS, 225 (78.7%) samples were positive for the mecA gene. There was 100% agreement between phenotypic identification and multiplex PCRs, similar to the present study (16). In a study by Montazeri et al, out of all the samples screened, 230 (53.11%) were Staphylococcus species (17). In a study by Nothando Florence Poyah, among 89 isolates obtained and identified as S. aureus, 48 (52.8%) had the mecA gene. In addition, regarding the presence of the Luks/f- pv gene, 16 isolates (17.97%) were reported as positive, of which 13 had the mecA gene or 3 lacked the mecA gene. Brown et al showed that more than 7% of the tested S. aureus strain samples had Luks/f- pv (18). Compared to the strains isolated from other parts of the country, the present research can be a suitable factor for investigating the geographical distribution of these strains. Further, infection control measures and compliance with health tips to eliminate or reduce these types of strains, especially in hospitals and intensive care units, should be taken very seriously. Considering the frequency of infections caused by staphylococcal species in society and the stability of these strains in nature due to their widespread resistance, the lack of using genotypic methods such as PCR to identify the virulence genes of these strains creates many problems, from Due to the fact that mecA gene requires special conditions for expression, as a result, the increase in mecA gene expression cannot be detected with usual laboratory tests. In genotyping tests, the presence of disease genes can be detected in the shortest time by using the multiplex PCR method with high sensitivity and specificity, and the creation of resistant strains can be prevented with this method. Conventional culture methods, selective and differential biochemical tests of media, and finally, serotyping are time-consuming and require live bacteria. Fast but sensitive diagnostic methods with high specificity are used in this regard (19). Currently, the development of molecular biology methods has facilitated the identification of biological agents in clinical samples with high accuracy and speed, among which PCR has a special place and has been suggested to confirm the pathogenic agent. One of the important methods with multiple efficiency is that by using the multiple PCR primer method, several bacteria can be simultaneously identified in a short period of time, which is a suitable method in terms of high sensitivity and specificity (20). Using phenotypic and serological methods, it is impossible to quickly identify the causative agents of staphylococci and eliminate the pathogenic agent (21).
Conclusion
Evaluating the prevalence of virulence gene strains can be effective in controlling infectious diseases in people with weakened immune systems and patients hospitalized in the intensive care unit. The multiplex PCR method for faster diagnosis of these genes can usually be performed in laboratories. Moreover, special measures to control infection, especially the frequency of virulence genes in the intensive care unit, can be an effective factor in reducing the pathogenicity of these genes.
Authors’ Contribution
Conceptualization: Babak Kheirkhah, Mitra Zare.
Data curation: Babak Kheirkhah, Iman Pouladi, Mitra Zare.
Formal analysis: Babak Kheirkhah, Iman Pouladi, Mitra Zare.
Investigation: Babak Kheirkhah, Mitra Zare.
Methodology: Babak Kheirkhah, Mitra Zare.
Project administration: Iman Pouladi, Babak Kheirkhah, Mitra Zare.
Resources: Iman Pouladi, Mitra Zare.
Writing–original draft: Iman Pouladi, Mitra Zare.
Writing–review & editing: Iman Pouladi, Mitra Zare.
Competing Interests
The authors declare that they have no conflict of interests.
Ethical Approval
This study is extracted from the Master’s thesis and has been approved by the Research Committee of Islamic Azad University, the Kerman branch, and an informed consent form was obtained from all participants.
Funding
None.
References
- Jauneikaite E, Ferguson T, Mosavie M, Fallowfield JL, Davey T, Thorpe N, et al. Staphylococcus aureus colonization and acquisition of skin and soft tissue infection among Royal Marines recruits: a prospective cohort study. Clin Microbiol Infect 2020;26(3):381.e1-381.e6. 10.1016/j.cmi.2019.07.014.
- Bukowski M, Wladyka B, Dubin G. Exfoliative toxins of Staphylococcus aureus. Toxins (Basel) 2010; 2(5):1148-65. doi: 10.3390/toxins2051148 [Crossref] [ Google Scholar]
- Khalili S, Bahrami N, Rezai S, Pouladi I. Prevalence of SEA and SEB enterotoxin producing methicillin-resistant Staphylococcus aureus strains among primary school children in Sari, Iran. Sri Lanka J Child Health 2021; 50(1):17-21. [ Google Scholar]
- Niakan M, Pouladi I, Kaviani R, Esmaili E. Antimicrobial effects of zinc oxide and silver nitrate nanoparticles on S aureus, A baumannii and P aeruginosa. J Basic Clin Pathophysiol 2019; 7(1):27-30. doi: 10.22070/jbcp.2019.3672.1105 [Crossref] [ Google Scholar]
- Ibberson CB, Whiteley M. The Staphylococcus aureus transcriptome during cystic fibrosis lung infection. mBio 2019; 10(6):e02774-19. doi: 10.1128/mBio.02774-19 [Crossref] [ Google Scholar]
- Tan X, Coureuil M, Ramond E, Euphrasie D, Dupuis M, Tros F. Chronic Staphylococcus aureus lung infection correlates with proteogenomic and metabolic adaptations leading to an increased intracellular persistence. Clin Infect Dis 2019; 69(11):1937-45. doi: 10.1093/cid/ciz106 [Crossref] [ Google Scholar]
- Stella AE, Lima TF, Moreira CN, De Paula EMN. Characterization of Staphylococcus aureus strains isolated from veterinary hospital. Int J Microbiol 2020; 2020:2893027. doi: 10.1155/2020/2893027 [Crossref] [ Google Scholar]
- Raheema RH, Abed KA. Molecular identification of virulence genes of Staphylococcus aureus isolated from different clinical isolates. Indian J Public Health Res Dev 2019; 10(2):212-8. [ Google Scholar]
- Mrochen DM, Schulz D, Fischer S, Jeske K, El Gohary H, Reil D. Wild rodents and shrews are natural hosts of Staphylococcus aureus. Int J Med Microbiol 2018; 308(6):590-7. doi: 10.1016/j.ijmm.2017.09.014 [Crossref] [ Google Scholar]
- Gonzalez AG, Marques LMP, da Silva Amorim Gomes M, do Couto Beltrão JC, Pinheiro MG, Esper LMR, et al. Methicillin-resistant Staphylococcus aureus in minas frescal cheese: evaluation of classic enterotoxin genes, antimicrobial resistance and clonal diversity. FEMS Microbiol Lett 2017;364(23). 10.1093/femsle/fnx232.
- Harrison EM, Paterson GK, Holden MT, Ba X, Rolo J, Morgan FJ. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J Antimicrob Chemother 2014; 69(4):911-8. doi: 10.1093/jac/dkt452 [Crossref] [ Google Scholar]
- Kaviani R, Pouladi I, Niakan M, Mirnejad R. Molecular detection of Adefg efflux pump genes and their contribution to antibiotic resistance in Acinetobacter baumannii clinical isolates. Rep Biochem Mol Biol 2020; 8(4):413-8. [ Google Scholar]
- Niakan M, Kaviani R, Heidari Motlagh R, Pouladi I. Investigation of the resistance of Acinetobactorbaumanii strains isolated from Tehran’s hospital inpatients in terms of selected antibiotics. J Basic Clin Pathophysiol 2018; 6(2):13-6. doi: 10.22070/jbcp.2018.3463.1101 [Crossref] [ Google Scholar]
- Beikmohammadi H, Viesy S, Kaviani R, Pouladi I. Detection of efflux pump genes conferring multidrug resistance in clinical isolates of Acinetobacter baumannii in Tehran province. Rev Res Med Microbiol 2022; 33(1):31-6. doi: 10.1097/mrm.0000000000000255 [Crossref] [ Google Scholar]
- Najafi Olya Z, Najar-Peerayeh S, Yadegar A, Bakhshi B. Clonal diversity and genomic characterization of Panton-Valentine leukocidin (PVL)-positive Staphylococcus aureus in Tehran, Iran. BMC Infect Dis 2021; 21(1):372. doi: 10.1186/s12879-021-06060-4 [Crossref] [ Google Scholar]
- Trevizani Rocchetti T, Benini Martins K, Faccioli-Martins PY, de Oliveira RA, Mondelli AL, Fortaleza C. Detection of the mecA gene and identification of Staphylococcus directly from blood culture bottles by multiplex polymerase chain reaction. Braz J Infect Dis 2018; 22(2):99-105. doi: 10.1016/j.bjid.2018.02.006 [Crossref] [ Google Scholar]
- Abbasi Montazeri E, Khosravi AD, Jolodar A, Ghaderpanah M, Azarpira S. Identification of methicillin-resistant Staphylococcus aureus (MRSA) strains isolated from burn patients by multiplex PCR. Burns 2015; 41(3):590-4. doi: 10.1016/j.burns.2014.08.018 [Crossref] [ Google Scholar]
- Poyah NF. Characterization of Methicillin Resistant Staphylococcus aureus Isolated from Raw Milk Obtained from the Eastern Cape Province. South Africa: University of Johannesburg; 2020.
- Aragão BB, Trajano SC, Silva JG, Silva BP, Oliveira RP, Junior JW. Short communication: high frequency of β-lactam-resistant Staphylococcus aureus in artisanal Coalho cheese made from goat milk produced in northeastern Brazil. J Dairy Sci 2019; 102(8):6923-7. doi: 10.3168/jds.2018-16162 [Crossref] [ Google Scholar]
- Kashani S, Alvandi AH, Abiri R. Diagnostic values of multiplex loop-mediated isothermal amplification and multiplex polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus. Jundishapur J Microbiol 2020; 13(6):e96682. doi: 10.5812/jjm.96682 [Crossref] [ Google Scholar]
- Khodabandeh M, Mohammadi M, Abdolsalehi MR, Alvandimanesh A, Gholami M, Bibalan MH. Analysis of resistance to macrolide-lincosamide-streptogramin B among mecA-positive Staphylococcus aureus isolates. Osong Public Health Res Perspect 2019; 10(1):25-31. doi: 10.24171/j.phrp.2019.10.1.06 [Crossref] [ Google Scholar]