Avicenna Journal of Clinical Microbiology and Infection. 10(3):88-94.
doi: 10.34172/ajcmi.3502
Original Article
Molecular Detection of Hospital-Acquired Methicillin-Resistant Staphylococcus aureus Isolated From Teaching Hospitals in Damascus, Syria
Lina ALyousef 1, *  , Salah Addin Shehadeh 1
, Salah Addin Shehadeh 1  , Ayman Al-Mariri 2
, Ayman Al-Mariri 2 
Author information:
1Department of Laboratory Medicine, Faculty of Medicine, Damascus University, Damascus, Syria
2Department of Molecular Biology and Biotechnology, Atomic Energy Commission of Syria, Damascus, Syria
Abstract
Background: Methicillin-resistant Staphylococcus aureus (MRSA) is an established pathogen responsible for hospital-acquired infections (HAIs). Accurate MRSA diagnosis is of paramount importance to facilitate early and effective treatment and to manage its transmission effectively. The primary objective of our study was to determine the precise prevalence of hospital-acquired MRSA (HA-MRSA) in select teaching hospitals in Damascus by detecting the presence of the mecA gene.
Methods: One hundred Staphylococcus aureus isolates were collected from various clinical specimens obtained from inpatients admitted to three major teaching hospitals in Damascus, Syria, including Al-Moussat, Al-Assad, and Tishreen Military Hospitals. These patients met the established criteria for HAIs. The isolates were collected between December 2021 and August 2022. Genus and species confirmation were conducted via polymerase chain reaction (PCR), employing the 16SrDNA gene specific to the Staphylococcus genus and the nuc gene specific to S. aureus. Methicillin resistance was assessed using cefoxitin disc diffusion (CDD) in accordance with Clinical and Laboratory Standards Institute (CLSI) recommendations. The presence of the mecA gene was also detected through PCR.
Results: Out of the collected isolates, 67% exhibited resistance to cefoxitin, as determined by the CDD, while 66% were found to be positive for the mecA gene. CDD demonstrated a sensitivity of 100% and a specificity of 97%.
Conclusion: This investigation revealed a notably high incidence of HA-MRSA infections within the teaching hospitals under scrutiny. The CDD method displayed significant sensitivity and specificity, making it a dependable alternative to the mecA PCR for MRSA detection. This finding holds substantial importance for the effective implementation of infection control initiatives and strategies aimed at eradicating MRSA and curtailing its spread within our hospital facilities.
Keywords: Methicillin-resistant Staphylococcus aureus, Hospital-acquired infections, Cefoxitin disc diffusion, mecA gene
Copyright and License Information
© 2023 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: ALyousef L, Shehadeh SA, Al-Mariri A. Molecular detection of hospital-acquired methicillin-resistant Staphylococcus aureus isolated from teaching hospitals in Damascus, Syria. Avicenna J Clin Microbiol Infect. 2023; 10(3):88-94. doi:10.34172/ajcmi.3502
Introduction
Staphylococcus aureus is one of the most prevalent pathogens found in healthcare facilities, causing a wide spectrum of infections in humans. Commonly affected areas include the skin and soft tissues, leading to conditions such as folliculitis, furuncles, and impetigo. Some of the diseases caused by this bacterium can be life-threatening, including bacteremia, endocarditis, pneumonia, and osteomyelitis, resulting in high rates of morbidity and mortality and a substantial economic burden (1).
The primary challenge posed by S. aureus is its remarkable resistance to various classes of antibiotics through various mechanisms. Within two years of the introduction of penicillin for the treatment of S. aureus infections, penicillin-resistant strains emerged due to the production of the beta-lactamase enzyme. However, just two years after the introduction of methicillin (which is a beta-lactamase-stable drug), methicillin-resistant S. aureus (MRSA) was first reported by the British scientist Jevons in 1961 (2). Currently, MRSA is regarded as a significant threat to human health, particularly in healthcare settings, as it exhibits resistance to all beta-lactam antibiotics and their derivatives, except for the latest generation. Moreover, some strains are resistant to other classes of antibiotics, such as quinolones, aminoglycosides, macrolides, and the like (3).
In accordance with the 2022 report from the Centers for Disease Control and Prevention (CDC), there has been a 7% increase in the incidence of MRSA infections within hospitals in the United States (4). Reports from Europe, Asia, and Africa have also substantiated the emergence of this resistant bacterium and underscored the pressing need for global intervention (5).
Methicillin resistance is primarily mediated by the mecA gene, which encodes novel penicillin-binding proteins (PBP2a, also known as PBP2′). There are additional, albeit less significant, mechanisms involving the overproduction of penicillinase or mutations causing alterations in natural PBPs (6). PBP2a exhibits low affinity for all beta-lactam drugs, rendering strains that express PBP2a resistant to penicillins, cephalosporins, monobactams, carbapenems, and beta-lactam drugs in conjunction with beta-lactamase inhibitors (6). MSSA strains, in contrast, possess natural PBPs and lack the mecA gene.
The CDC defines healthcare-associated infections as localized or systemic conditions resulting from an adverse reaction to the presence of infectious agents or their toxins that were not present upon admission to the healthcare facility. Conventionally, a 48-hour time threshold after admission is employed to distinguish between hospital-acquired and community-acquired infections (7).
The prevalence of hospital-acquired infections (HAIs) in developed nations stands at approximately 5%–10%, which is rising to 20%–25% in developing countries (8). The incidence of HA-MRSA strains varies from one country to another due to various factors, including antibiotic treatment policies, the implementation of infection control programs, and the rate of nasal carriers. Additionally, this prevalence is on the rise worldwide, owing to the emergence of new evolutionary MRSA strains that demonstrate greater adaptability to the hospital environment (9).
Similar to many developing countries, Syria is grappling with a shortage of medical capabilities and a decline in infection control and epidemiological investigation protocols, exacerbated by the conditions of conflict and war. The formulation of an effective policy to mitigate the incidence of nosocomial infections caused by HA-MRSA hinges on acquiring precise insights into their prevalence and epidemiology.
To date, there have been no local studies conducted in Syrian hospitals that establish the exact frequency of MRSA, except for studies targeting specific groups, such as medical staff (10), febrile neutropenic patients (11), or Syrian refugees (12). However, these studies have predominantly relied on conventional diagnostic methods for identifying MRSA.
To the best of our knowledge, this study marks the first instance of employing molecular techniques, specifically the polymerase chain reaction (PCR) method, to ascertain the precise prevalence of MRSA. The detection of MRSA is facilitated through the examination of the mecA gene, a widely acknowledged gold standard in MRSA identification.
Materials and Methods
Clinical Sampling and Laboratory Identification
Overall, 100 non-duplicative clinical samples were collected from the microbiology laboratories of three teaching hospitals in Damascus (Al-Mouassat Hospital, Al-Assad Hospital, and Tishreen Military Hospitals) between December 2021 and August 2022. After reviewing the medical file of each patient, only isolates that meet the criteria for HAIs according to the definition of the CDC were chosen for the study. The practical part of the study was conducted in the laboratory of the Department of Molecular Biology and Biotechnology in the Atomic Energy Commission of Syria (AECS). Standard strains of MRSA (ATCC® 43300) and MSSA (ATCC® 25923) were used as control samples in this study.
Laboratory Identification
Gram-positive cocci appeared in clusters, catalase-positive, mannitol fermenter, and coagulase-positive bacteria, were initially identified as S. aureus; then, the identification was confirmed based on the presence of the 16S rDANand nuc genes, specific to the Staphylococcus genusand S. aureus, respectively, using the PCR technique. Primers were designed for the studied genes using specialized software (Geneious). The specific sequences of the primers utilized in the molecular identification are provided in Table 1. Bacterial DNA was extracted using the cetyl trimethyl ammonium bromide (CTAB) method (13) and then stored at -20 °C.
Table 1.
Sequence of Primers
|
Recognized Gene
|
Sequence of Primer 5'-3'
|
Size (bp)
|
|
16S rDNA
|
GGAATTCAAAKGAATTGACGGGGGC |
993 |
| CGGGATCCCAGGCCCGGGAACGTATTCAC |
|
nuc
|
GCGATTGATGGTGATACGGTT |
270 |
| AGCCAAGCCTTGACGAACTAAAGC |
|
mecA
|
TGCTATCCACCCTCAAACAGG |
286 |
| AACGTTGTAACCACCCCAAGA |
The PCR mixture included 1 µL extracted DNA (100 ng), 1 µL of each primer (10 µM), 1 µL (10 µM) of dNTPs (Fermentas, USA), 5 µL of 10 × buffer (Fermentas, USA), 3 µL of MgSO4 (50 mg/mL), and 1 µL (5U) taq polymerase (Vivantis, Malaysia), and the final volume was completed to 50 µL with nuclease-free water. The PCR was performed in the PCR Thermal Cycler (TECHNE, USA). The conditions of the reaction are presented in Table 2. PCR products were electrophoretically separated in 1% agarose gel (Sigma, USA) containing 1 µg/µL ethidium bromide and visualized under a UV transilluminator (UV tec, Korea).
Table 2.
The PCR Program
|
Steps
|
Temperature
|
Time
|
Cycles
|
| Initial denaturation |
95 ° |
5 min |
|
| Denaturation |
95 ° |
30 s |
35 cycles |
| Annealing |
58 ° |
45 s |
| Elongation |
72 ° |
1 min |
| Final extension |
72 ° |
10 min |
|
| Hold |
4 ° |
Note. PCR: Polymerase chain reaction.
Cefoxitin Sensitivity Test
In general, 100 isolates were tested for their sensitivity to cefoxitin disc 30 μg (Condalab, Spain) by the disc diffusion method CDD according to the recommendation of the Clinical and Laboratory Standards Institute (CLSI) (14).
In brief, a colony from an overnight Luria-Bertani (LB) agar plate was transferred to a tube of 5 mL of LB broth. The turbidity was adjusted to 0.5 McFarland equal to 1.5 × 108 CFU/mL, and the bacterial suspension was inoculated on a Muller-Hinton agar plate of 9 cm in diameter. The plates were left to dry for 10–15 minutes. The disc of cefoxitin (30 µg) was put in the middle of each inoculated plate. The plates were then incubated at 30 °C for 16–18 hours.
The results were read by measuring the diameter of the inhibition halo around the disc. The isolate was classified as resistant (R) if the diameter of the inhibition zone was ≤ 21 mm and sensitive (S) if the diameter was ≥ 22 mm, according to CLSI guidelines (14).
mecA Polymerase Chain Reaction
The extracted bacterial DNA according to the CTAB method was used (13). Primers (Table 1) were designed for the mecA gene using specialized software (Geneious).
PCR was performed in the final volume of 25 μL. The master mix composition was 2.5 µL of 10 × buffer (Fermentas, USA), 1.5 µL (50 mg/mL) of MgSo4, 1 µL (10 µM) of dNTPs (Fermentas, USA), 2 µL of each primer (10 µM), 0.5 μL (5 U) Taq polymerase (Vivantis, Malaysia), and 2 µL (100 ng) of extracted DNA. The volume was completed to 25 µL with sterile nuclease-free distilled water. The positive controls were obtained from AECS, and a PCR master mix without a DNA template was used as a negative control. PCR was conducted using a thermal cycler (TECHNE, USA), and the amplification cycle conditions are summarized in Table 2.
PCR products were electrophoretically separated in 1% agarose gel (Sigma, USA) containing 1 µg/µL ethidium bromide and visualized under a UV transilluminator (UV tec, Korea).
Statistical Analysis
Descriptive statistical analysis, including percentages to characterize data, was performed using Microsoft Excel 2010 (Microsoft Corp; Redmond, WA, USA).
The sensitivity and specificity of the cefoxitin disc diffusion method (CDD) were calculated using mecA gene PCR as the gold standard.
Results
Characterization of Samples According to Gender and Age
Age of the patients (62 males and 38 females) included in the study ranged between 2 and 88 years, with an average age of 47 years.
Results of Molecular Identification Using 16S rDAN and nuc Genes
One hundred isolates were confirmed to be S. aureus using 16S rDANand nuc gene PCR. A part of the results is illustrated in Figures 1 and 2.
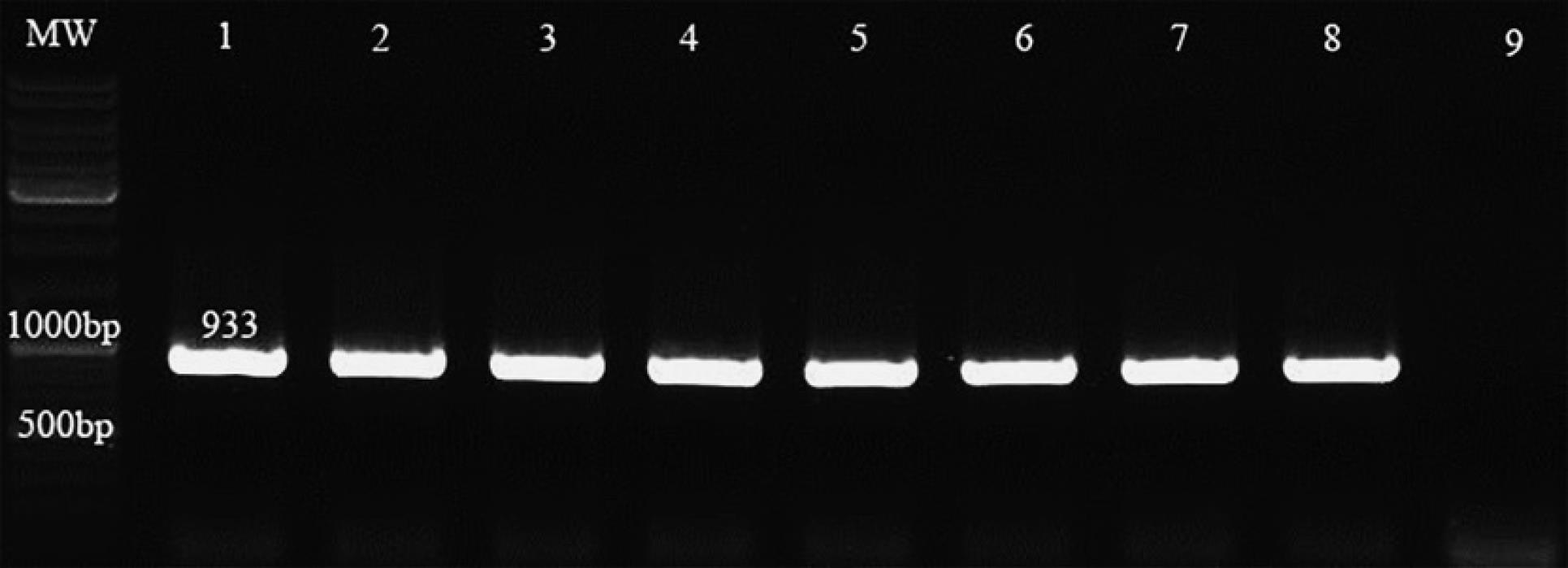
Figure 1.
Results of 16S rDNA Polymerase Chain Reaction. Note. PCR: Polymerase chain reaction; MW: DNA leader -Lane (1) Control positive- Lane (9) Control negative- Lanes (2-8) isolates carry the 16S rDNA gene.
.
Results of 16S rDNA Polymerase Chain Reaction. Note. PCR: Polymerase chain reaction; MW: DNA leader -Lane (1) Control positive- Lane (9) Control negative- Lanes (2-8) isolates carry the 16S rDNA gene.
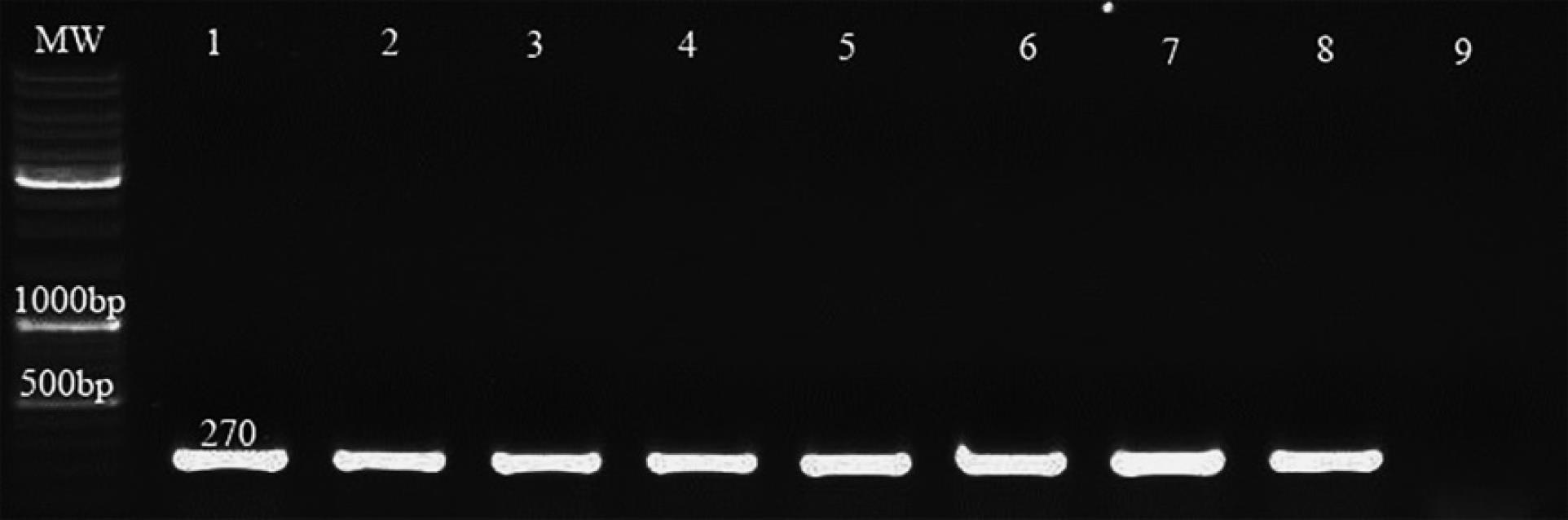
Figure 2.
Results of nuc Polymerase Chain Reaction. Note. PCR: Polymerase chain reaction; MW: DNA leader –Lane (1) Control positive- Lane (9) Control negative- Lanes (2,3,4,5,6,7,8) isolates carry the nuc gene
.
Results of nuc Polymerase Chain Reaction. Note. PCR: Polymerase chain reaction; MW: DNA leader –Lane (1) Control positive- Lane (9) Control negative- Lanes (2,3,4,5,6,7,8) isolates carry the nuc gene
Results of Cefoxitin Disc Diffusion Test
By the disc-diffusion method (Kirby-Bauer), 100 S. aureus isolates were tested for their sensitivity to cefoxitin (30 µg), and 67 (67%) and 33 (33%) isolates showed resistance to and were sensitive to cefoxitin, respectively.
Results of mecA Polymerase Chain Reaction
Sixty-six isolates were positive for the mecA gene according to PCR, with a rate of 66%, and were classified as MRSA, and thirty-four (34%) were negative for this gene and classified as methicillin-susceptible S. aureus (MSSA). All isolates that were resistant to cefoxitin by the disc diffusion test possessed the mecA gene, except for one isolate, which was negative for the mecA gene. A part of the results of mecA PCR is depicted in Figure 3. The percentages of MRSA and MSSA are illustrated in Figure 4.

Figure 3.
Results of mecA Gene Screening by Polymerase Chain Reaction. Note. Isolates (M13-M31-M43-T40-T43) are mecA gene positive
.
Results of mecA Gene Screening by Polymerase Chain Reaction. Note. Isolates (M13-M31-M43-T40-T43) are mecA gene positive
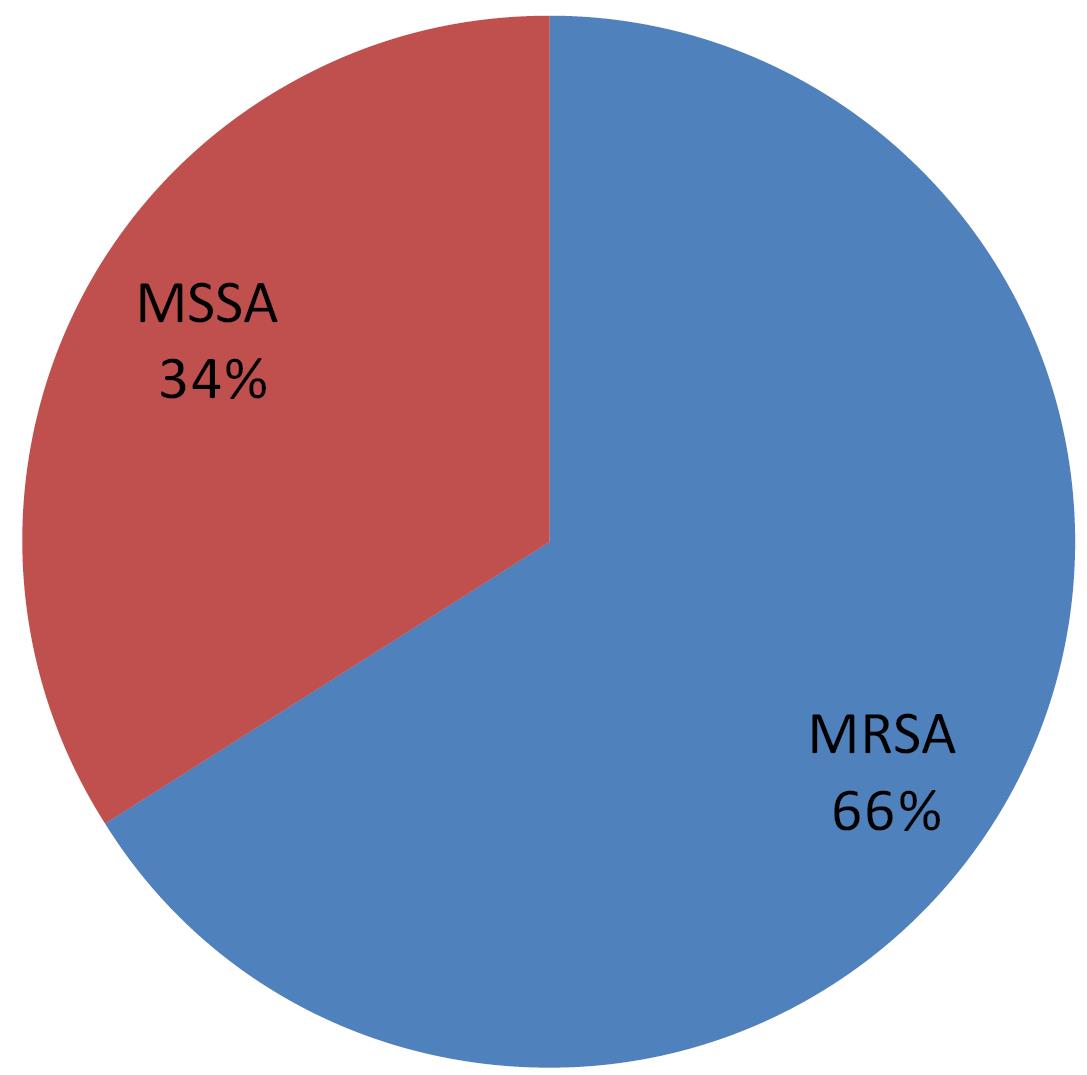
Figure 4.
MRSA Percentage According to mecA Polymerase Chain Reaction. Note. MRSA: Methicillin-resistant Staphylococcus aureus; PCR: Polymerase chain reaction
.
MRSA Percentage According to mecA Polymerase Chain Reaction. Note. MRSA: Methicillin-resistant Staphylococcus aureus; PCR: Polymerase chain reaction
Depending on the fact that mecA gene positivity is the gold standard in diagnosing MRSA, the sensitivity and specificity of CDD were calculated as follows:
The sensitivity of CDD = True positive (66) / True positive (66) + False negative (0) = 100%
The specificity of CDD = True negative (34) / True negative (34) + False positive (1) = 97%
Distribution of Samples According to Type
Among fifty-seven samples of pus, forty samples (70.2%) were positive for the mecA gene and classified as MRSA. MRSA consisted of 57.1% (8/14) of blood samples, 70% (7/10) of sputum samples, 57.1% (8/14) of urine samples, and 60% (3/5) of other types of samples. The highest percentage of MRSA was from pus and sputum (Table 3).
Table 3.
Types of MRSA Samples
|
Sample Type
|
No. of
Staphylococcus aureus
|
No. of MRSA
|
MRSA (%)
|
| Pus |
57 |
40 |
70.2 |
| Blood |
14 |
8 |
57.1 |
| Sputum |
10 |
7 |
70 |
| Urine |
14 |
8 |
57.1 |
| others |
5 |
3 |
60 |
Note. MRSA: Methicillin-resistant Staphylococcus aureus.
Distribution of Samples According to the Ward
Out of nine isolates taken from the burn unit, eight isolates (8/9, 88.9%) were positive for the mecA gene and considered MRSA. MRSA consisted of 76.9% (10/13), 69.1% (38/55), and 34.5% (10/23) of isolates taken from the intensive care unit (ICU), surgery wards, and internal wards, respectively (Table 4).
Table 4.
Distribution of MRSA in the Hospital Wards
|
Ward
|
No. of
Staphylococcus aureus
|
No. of MRSA
|
MRSA (%)
|
| Surgery wards |
55 |
38 |
69.1 |
| Internal wards |
23 |
10 |
34.5 |
| ICU |
13 |
10 |
76.9 |
| Burn unit |
9 |
8 |
88.9 |
Note. MRSA: Methicillin-resistant Staphylococcus aureus; ICU: Intensive care unit.
Discussion
The findings demonstrated a significant prevalence of methicillin resistance in the strains of S. aureus responsible for HAIs (HA-MRSA) in our teaching hospitals, with a recorded rate of 66% (Figure 4). This percentage exceeds the findings of a previous local study, which reported a rate of 33.3% (15). However, it is worth noting that the aforementioned study did not distinguish between hospital-acquired and community-acquired infections. In another local study, MRSA was found at a rate of 9.4%, albeit in a study group consisting of nasal carriers among medical staff (10). A recent study at Al-Mouassat hospital reported a 5.4% incidence of MRSA, but this study was confined to patients with febrile neutropenia (11).
Our study is pioneering in its investigation of HA-MRSA infections using molecular techniques for detecting the mecA gene, widely recognized as the gold standard for MRSA diagnosis. In comparison to international studies, our percentage aligns with Iranian studies, which reported rates ranging from 40% to 57% (16). An Indian study recorded a 70% prevalence in this regard (17). Even in a study in the United States, the percentage of methicillin-resistant strains among S. aureus causing septicemia was 48.5% (18).
These elevated rates continue to be causes of concern for health authorities worldwide. They impose additional burdens on medical staff and patients, leading to extended hospitalization periods and heightened demand for supplementary treatments, ultimately contributing to increased morbidity and mortality rates.
The substantial percentage of HA-MRSA in our study may be attributed to various factors, including insufficient adherence to prevention and infection control programs, the inappropriate use and overuse of antibiotics, and the absence of surveillance programs to monitor asymptomatic carriers, whether among medical staff or admitted patients. Multiple studies affirm that the rigorous implementation of prophylactic hygiene standards and comprehensive staff training play pivotal roles in controlling the spread of MRSA in hospital settings (19).
The highest rates of MRSA were found in pus and sputum (70.2% and 70%, Table 3). It is important to note that S. aureus, including MRSA, is part of the normal microbiome of the skin and pharynx in some individuals (20), with global studies estimating that approximately 30% of the population are permanent asymptomatic carriers. In such cases, due to the immunodeficient state associated with hospitalization, these bacteria can trigger endogenous infections in their respective niches (21,22). Consequently, several international studies have recommended swabbing the nose or pharynx of newly admitted patients to investigate whether they are asymptomatic carriers of MRSA (23,24).
In our study, MRSA was prevalent in the burn unit at a high rate of 88.9%, followed by the ICU at 76.9%, surgical wards at 69.1%, and the lowest incidence at 34.5% was observed in various internal wards (Table 4). These findings align with results from numerous studies conducted in other countries (22,25). This can be attributed to burn patients having lost their natural skin barrier, which serves as the first line of defense against infections, rendering them susceptible to skin infections by bacteria such as MRSA. Patients in surgical wards are exposed to a range of surgical procedures, including the use of intravenous and urinary catheters or implants (26). ICU patients, given their immunocompromised status and the use of mechanical ventilation and invasive procedures, are particularly at risk of HAIs (27,28).
Invasive medical procedures and the use of implants increase susceptibility to HAIs (9). Some MRSA strains exhibit a strong ability to colonize surfaces and form biofilms, particularly on intravenous and urinary catheters or bone implants (29). Biofilms allow bacteria to resist antibiotic treatment at conventional doses and provide protection against the immune system. These biofilm-coated tools become a source of bacterial contamination. Periodic monitoring and replacement of catheters are recommended in line with international guidelines (30).
Our findings demonstrated a strong correlation between sensitivity to cefoxitin using the disc-diffusion method and the detection of the mecA gene through PCR. The sensitivity and specificity were 100% and 97%, respectively, in line with several studies, suggesting that resistance to cefoxitin serves as a robust indicator of the presence of the mecA gene (31). However, it is important to note that this method may fail to distinguish certain strains that possess mecA but still exhibit sensitivity to cefoxitin using the disc-diffusion technique. This phenomenon is attributed to the instability of the gene (32), although such isolates were not observed in this study. On the contrary, an isolate was detected that displayed resistance to cefoxitin using the disc-diffusion method, but mecA detection was negative, which is a scenario reported in various studies (33,34). This discrepancy can be attributed to factors such as hyperproduction of penicillinase, mutations in normal penicillin-binding proteins, or the presence of other resistance-mediating genes such as mecC, mecB, or mecD, encoding alternative penicillin-binding proteins sharing amino acid sequences with PBP2a ranging from 51% to 63% (2). In such cases, the disc diffusion method is preferred over molecular techniques, as it classifies such isolates as MRSA. To address this gap in molecular methods, it is advisable to incorporate primers for mecC, mecB, or mecD into PCR, particularly since these strains are often associated with animal sources and are known as livestock-associated MRSA, with a noticeable increase in prevalence (2).
The laboratories of the teaching hospitals in Damascus, included in this study, opt for the disc-diffusion method for MRSA detection due to the lack of reagents for automated devices and the high workload that does not align with the application of molecular methods. It is important to note that international recommendations favor the use of cefoxitin over oxacillin due to its greater stability against penicillinase. Therefore, it is imperative to disseminate and publish unified and regularly updated protocols in accordance with the latest international guidelines to enhance the accuracy of MRSA diagnosis in our laboratories. The disc diffusion method, being straightforward, cost-effective, and not reliant on complex conditions, is preferred by numerous studies over other methods (35).
Conclusion
The findings of this study revealed a high frequency of HA-MRSA among the clinical S. aureus isolates from the inpatients of Syrian teaching hospitals. This situation requires the development of strict programs to track these bacteria, identify their reservoirs, and emphasize applying strict measures to control their infections and prevent their spread, especially in units with a high frequency of MRSA infections, such as burn units and ICUs. The CDD method displays a reliable alternative to mecA PCR to detect MRSA and can be used routinely in resource-limited settings. The emphasis must be placed on hygienic handwashing and all necessary sterilization and disinfection procedures before and after dealing with patients. Such procedures have succeeded in many countries in reducing the rate of HAIs caused by MRSA, and this reduces the moral and financial burden that these infections cause to patients and health authorities.
Acknowledgements
We are grateful to the staff of theDepartment of Molecular Biology and Biotechnology in the Atomic Energy Commission of Syria for their cooperation.
Authors’ Contribution
Conceptualization: Lina ALyousef, Salah Addin Shehadeh, Ayman Al-Mariri.
Data curation: Lina ALyousef, Ayman Al-Mariri.
Formal analysis: Lina ALyousef.
Investigation: Lina ALyousef, Salah Addin Shehadeh.
Methodology: Lina ALyousef, Salah Addin Shehadeh, Ayman Al-Mariri.
Project administration: Lina ALyousef, Salah Addin Shehadeh, Ayman Al-Mariri.
Resources: Lina ALyousef, Salah Addin Shehadeh, Ayman Al-Mariri.
Supervision: Lina ALyousef, Salah Addin Shehadeh, Ayman Al-Mariri.
Validation: Lina ALyousef, Salah Addin Shehadeh, Ayman Al-Mariri.
Visualization: Lina ALyousef, Ayman Al-Mariri.
Writing–original draft: Lina ALyousef.
Writing–review & editing: Lina ALyousef.
Competing Interests
The authors declare no conflict of interest.
Ethical Approval
The Ethics Committee of Damascus University approved the research protocol.
Funding
This research was funded by Damascus University (Funder No. 501100020595).
References
- Becker A, Forster DH, Kniehl E. Oxacillin resistance screening agar base for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2002; 40(11):4400-1. doi: 10.1128/jcm.40.11.4400-4401.2002 [Crossref] [ Google Scholar]
- Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 2018; 31(4):e00020-18. doi: 10.1128/cmr.00020-18 [Crossref] [ Google Scholar]
- Mlynarczyk-Bonikowska B, Kowalewski C, Krolak-Ulinska A, Marusza W. Molecular mechanisms of drug resistance in Staphylococcus aureus. Int J Mol Sci 2022; 23(15):8088. doi: 10.3390/ijms23158088 [Crossref] [ Google Scholar]
- Baker MA, Sands KE, Huang SS, Kleinman K, Septimus EJ, Varma N. The impact of coronavirus disease 2019 (COVID-19) on healthcare-associated infections. Clin Infect Dis 2022; 74(10):1748-54. doi: 10.1093/cid/ciab688 [Crossref] [ Google Scholar]
- Borg MA, Camilleri L. What is driving the epidemiology of methicillin-resistant Staphylococcus aureus infections in Europe?. Microb Drug Resist 2021; 27(7):889-94. doi: 10.1089/mdr.2020.0259 [Crossref] [ Google Scholar]
- Palavecino EL. Clinical, epidemiologic, and laboratory aspects of methicillin-resistant Staphylococcus aureus infections. In: Ji Y, ed. Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols: Cutting-Edge Technologies and Advancements. New York, NY: Springer US; 2020. p. 1-28. 10.1007/978-1-4939-9849-4_1.
- Kouchak F, Askarian M. Nosocomial infections: the definition criteria. Iran J Med Sci 2012; 37(2):72-3. [ Google Scholar]
- Li Y, Gong Z, Lu Y, Hu G, Cai R, Chen Z. Impact of nosocomial infections surveillance on nosocomial infection rates: a systematic review. Int J Surg 2017; 42:164-9. doi: 10.1016/j.ijsu.2017.04.065 [Crossref] [ Google Scholar]
- Friedrich AW. Control of hospital acquired infections and antimicrobial resistance in Europe: the way to go. Wien Med Wochenschr 2019; 169(Suppl 1):25-30. doi: 10.1007/s10354-018-0676-5 [Crossref] [ Google Scholar]
- Tabana YM, Dahham S, Al-Hindi B, Al-Akkad A, Khadeer Ahamed MB. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) among medical staff in three Syrian provinces: Damascus, Daraa and Al-Swayda. Middle East J Sci Res 2015; 23(8):1756-64. doi: 10.5829/idosi.mejsr.2015.23.08.12633 [Crossref] [ Google Scholar]
- Alrstom A, Daher N, Abouharb R. Bloodstream infection antibiogram in Syrian febrile neutropenic patients. Int J Infect Dis 2020; 101(Suppl 1):26. doi: 10.1016/j.ijid.2020.09.104 [Crossref] [ Google Scholar]
- Rasheed NA, Hussein NR. Methicillin-resistant Staphylococcus aureus carriage rate and molecular characterization of the staphylococcal cassette chromosome mec among Syrian refugees in Iraq. Int J Infect Dis 2020; 91:218-22. doi: 10.1016/j.ijid.2019.12.006 [Crossref] [ Google Scholar]
- Ashwini D, Tiwari SP. Use of CTAB method for isolation of good quality and quantity of DNA. J Pure Appl Microbiol 2015; 9(3):2271-4. [ Google Scholar]
- Humphries R, Bobenchik AM, Hindler JA, Schuetz AN. Overview of changes to the Clinical and Laboratory Standards Institute performance standards for antimicrobial susceptibility testing, M100, 31st edition. J Clin Microbiol 2021; 59(12):e0021321. doi: 10.1128/jcm.00213-21 [Crossref] [ Google Scholar]
- Ali R, Al-Achkar K, Al-Mariri A, Safi M. Role of polymerase chain reaction (PCR) in the detection of antibiotic-resistant Staphylococcus aureus. Egypt J Med Hum Genet 2014; 15(3):293-8. [ Google Scholar]
- Ahmadishoar S, Kazemipour N, Sadeghi J, Nahaei MR, Kheirkhah B. Genotypic and phenotypic characterisation of clinical isolates of methicillin-resistant Staphylococcus aureus in two different geographical locations of Iran. Indian J Med Microbiol 2020; 38(2):162-8. doi: 10.4103/ijmm.IJMM_20_153 [Crossref] [ Google Scholar]
- Rasool MS, Ajaz M, Shar AH, Rind NA, Qurat-Ul-Ain Qurat-Ul-Ain, Shaikh F. Molecular characterization of drug resistant indigenous methicillin-resistant Staphylococcus aureus (MRSA) strains. Acta Biomed 2023; 94(6):e2023072. doi: 10.23750/abm.v94iS1.14682 [Crossref] [ Google Scholar]
- Inagaki K, Lucar J, Blackshear C, Hobbs CV. Methicillin-susceptible and methicillin-resistant Staphylococcus aureus bacteremia: nationwide estimates of 30-day readmission, in-hospital mortality, length of stay, and cost in the United States. Clin Infect Dis 2019; 69(12):2112-8. doi: 10.1093/cid/ciz123 [Crossref] [ Google Scholar]
- Sadeghi Moghaddam T, Namaei MH, Afshar D, Yousefi M. High frequency of SCCmec type IV and multidrug-resistant SCCmec type I among hospital acquired methicillin-resistant Staphylococcus aureus isolates in Birjand Imam Reza hospital, Iran. Iran J Microbiol 2022; 14(1):67-75. doi: 10.18502/ijm.v14i1.8803 [Crossref] [ Google Scholar]
- Mehrshad S, Haghkhah M, Aghaei S. Epidemiology and molecular characteristics of methicillin-resistant Staphylococcus aureus from skin and soft tissue infections in Shiraz, Iran. Turk J Med Sci 2017; 47(1):180-7. doi: 10.3906/sag-1507-164 [Crossref] [ Google Scholar]
- Ali T, Basit A, Karim AM, Lee JH, Jeon JH, Rehman SU. Mutation-based antibiotic resistance mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. Pharmaceuticals (Basel) 2021; 14(5):420. doi: 10.3390/ph14050420 [Crossref] [ Google Scholar]
- Schweizer M, Ward M, Cobb S, McDanel J, Leder L, Wibbenmeyer L. The epidemiology of methicillin-resistant Staphylococcus aureus on a burn trauma unit. Infect Control Hosp Epidemiol 2012; 33(11):1118-25. doi: 10.1086/668032 [Crossref] [ Google Scholar]
- Nakamura M, Shimakawa T, Nakano S, Chikawa T, Yoshioka S, Kashima M. Screening for nasal carriage of Staphylococcus aureus among patients scheduled to undergo orthopedic surgery: incidence of surgical site infection by nasal carriage. J Orthop Sci 2017; 22(4):778-82. doi: 10.1016/j.jos.2017.03.005 [Crossref] [ Google Scholar]
- Borg MA, Suda D, Scicluna E, Brincat A, Zarb P. Universal admission screening: a potential game-changer in hospitals with high prevalence of MRSA. J Hosp Infect 2021; 113:77-84. doi: 10.1016/j.jhin.2021.03.024 [Crossref] [ Google Scholar]
- Kalligeros M, Shehadeh F, Karageorgos SA, Zacharioudakis IM, Mylonakis E. MRSA colonization and acquisition in the burn unit: a systematic review and meta-analysis. Burns 2019; 45(7):1528-36. doi: 10.1016/j.burns.2019.05.014 [Crossref] [ Google Scholar]
- Kejela T, Dekosa F. High prevalence of MRSA and VRSA among inpatients of Mettu Karl Referral Hospital, Southwest Ethiopia. Trop Med Int Health 2022; 27(8):735-41. doi: 10.1111/tmi.13789 [Crossref] [ Google Scholar]
- Lin MY, Hayden MK, Lyles RD, Lolans K, Fogg LF, Kallen AJ. Regional epidemiology of methicillin-resistant Staphylococcus aureus among adult intensive care unit patients following state-mandated active surveillance. Clin Infect Dis 2018; 66(10):1535-9. doi: 10.1093/cid/cix1056 [Crossref] [ Google Scholar]
- Alp E, Yerer M, Esel D, Metan G, Doğanay M. Risk factors for acquisition of methicillin-resistant Staphylococcus aureus and clonal spread of the isolates in a medical intensive care unit. Turk J Med Sci 2009; 39(6):941-51. doi: 10.3906/sag-0801-7 [Crossref] [ Google Scholar]
- Silva V, Almeida L, Gaio V, Cerca N, Manageiro V, Caniça M. Biofilm formation of multidrug-resistant MRSA strains isolated from different types of human infections. Pathogens 2021; 10(8):970. doi: 10.3390/pathogens10080970 [Crossref] [ Google Scholar]
- Sohail M, Latif Z. Molecular analysis, biofilm formation, and susceptibility of methicillin-resistant Staphylococcus aureus strains causing community- and health care-associated infections in central venous catheters. Rev Soc Bras Med Trop 2018; 51(5):603-9. doi: 10.1590/0037-8682-0373-2017 [Crossref] [ Google Scholar]
- Rajeswarie S, Pradha V, D’Souza AO, Vinod R. Comparison of phenotypic and genotypic characterization methods for the detection of methicillin-resistant Staphylococcus aureus. Cureus 2022; 14(3):e23396. doi: 10.7759/cureus.23396 [Crossref] [ Google Scholar]
- Gargis AS, Yoo BB, Lonsway DR, Anderson K, Campbell D, Ewing TO. Difficult-to-detect Staphylococcus aureus: mecA-positive isolates associated with oxacillin and cefoxitin false-susceptible results. J Clin Microbiol 2020; 58(4):e02038-19. doi: 10.1128/jcm.02038-19 [Crossref] [ Google Scholar]
- Olayinka BO, Olayinka AT, Obajuluwa AF, Onaolapo JA, Olurinola PF. Absence of mecA gene in methicillin-resistant Staphylococcus aureus isolates. Afr J Infect Dis 2009; 3(2):49-56. doi: 10.4314/ajid.v3i2.55081 [Crossref] [ Google Scholar]
- Ba X, Harrison EM, Edwards GF, Holden MT, Larsen AR, Petersen A. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin-resistant on susceptibility testing, but lack the mec gene. J Antimicrob Chemother 2014; 69(3):594-7. doi: 10.1093/jac/dkt418 [Crossref] [ Google Scholar]
- Kumar A, Kashyap B, Mehndiratta A. Comparison of conventional and rapid methods for the detection of methicillin-resistance in Staphylococcus aureus from blood cultures. Trop J Med Res 2017; 20(2):196-200. doi: 10.4103/tjmr.tjmr_48_16 [Crossref] [ Google Scholar]