Avicenna Journal of Clinical Microbiology and Infection. 10(3):126-130.
doi: 10.34172/ajcmi.3460
Original Article
Molecular Detection of Microsporidia in Cattle in Jahrom, Iran
Amin Shafiee 1  , Gholamreza Shokoohi 1, Aminallah Saadatnia 1, Ahmad Abolghazi 2, *
, Gholamreza Shokoohi 1, Aminallah Saadatnia 1, Ahmad Abolghazi 2, * 
Author information:
1Department of Medical Parasitology, Jahrom University of Medical Sciences, Jahrom, Iran
2Department of Medical Parasitology, Hamadan University of Medical Sciences, Hamadan, Iran
Abstract
Background: Microsporidia are eukaryotic, single-celled intracellular parasites that can produce spores. Recently, they have been considered one of the opportunistic pathogens causing chronic diseases. There are more than 140 genera and 1200 species of microsporidia. Many human microsporidia are probably of zoonotic origin and are transmitted by contaminated water with animal feces. Given the zoonotic importance of this parasite and its ability to be transmitted from animals to humans, diagnosing and determining the species of parasite would seem essential for health strategies.
Methods: Two hundred fecal samples of slaughtered cows were collected from the Jahrom abattoirs from February 2021 to January 2022 and examined by molecular methods, including polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP-PCR).
Results: From the 200 samples examined by PCR, 19 (9.5%) samples tested positive for microsporidia, of which 17 isolates were Enterocytozoon bieneusi and two isolates were Encephalitozoon cuniculi.
Conclusion: The results revealed that microsporidia were present in cow feces. In addition, these findings indicated that cows can be considered a source of contamination for microsporidia. Given that this disease is a zoonosis, it is highly important to pay attention to the presence of this parasite in domestic animals that are in contact with humans. Further studies must be performed in different regions and different animals to understand the epizootiology of the pathogen. Eventually, the wide host range of microsporidia necessitates accurate identification of species and genera in all hosts all over the world.
Keywords: Microsporidia, Abattoirs, Cattle, Jahrom, PCR-RFLP
Copyright and License Information
© 2023 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Shafiee A, Shokoohi G, Saadatnia A, Abolghazi A. Molecular detection of microsporidia in cattle in Jahrom, Iran. Avicenna J Clin Microbiol Infect. 2023; 10(3):126-130. doi:10.34172/ajcmi.3460
Introduction
Microsporidiosis is an infectious disease caused by protozoan parasites belonging to the phylum microsporidia. These parasites have the ability to infect vertebrates and invertebrates (1-3). Microsporidia are intracellular parasites consisting of 140 genera and 1200 species of microsporidia (4). Two genera, Encephalitozoon and Enterocytozoon, have been reported frequently from a wide range of mammalian hosts (5,6). They infect the intestinal, ocular, and muscular tissues of mammals (6). These parasites have been reported from all over the world and are now recognized as one of the most common pathogens in patients with immune deficiency syndrome (7,8). In Iran, studies in Khuzestan province have reported the highest prevalence (26.5%) of animal microsporidia (9). In addition, studies in Iran have demonstrated that the frequency of microsporidia is approximately 6% (10). However, in two studies from Lorestan and Bushehr, the rate of microsporidia infection was reported to be zero (10,11). This parasite has been reported in the feces of animals such as donkeys, dogs, pigs, cows, and goats (6,12,13). Clinical signs depend on the species and immune status of the host. Many human microsporidia are probably of zoonotic origin and are transmitted by water contaminated with animal feces (13,14). Cows can be infected with microsporidia due to their close contact with humans. Thus, determining its prevalence in each region is important (15). The objective of this study was to determine the frequency of microsporidia in cattle feces in Jahrom. In this study, the molecular method was used to detect microsporidia spores in the fecal samples of cows from Jahrom (Fars, Iran). The survey was designed to expand our knowledge of the pathogenic role of microsporidia in animals having close contact with humans.
Materials and Methods
Study Area
Jahrom is one of the cities of Fars province, located in the southern part of the region. The town is 5436 km2 in area. This city is located within longitudes of 52° 45′ to 54° 4′ E and latitudes of 28° 19′ to 29° 10′ N (16).
Animal Stool Samples
A total of 200 fecal samples were collected (each sample about 100 g) from slaughtered cows (All cows were Holstein Friesians, and the average age of the cows was 2 years) in the abattoirs of Jahrom and immediately transferred to the Parasitology Laboratory of Jahrom University of Medical Sciences under sterile conditions from February 2021 to January 2022. When the cows were slaughtered, an incision was made in the cow’s colon with a scalpel, and in completely sterile conditions, the samples were collected and stored at 4 °C until DNA extraction (17).
DNA Extraction
DNA was extracted from all samples according to the instructions of Bioneer Company’s DNA extraction kit. Purified DNA is kept at -20 °C until it is used. The targets for the nested polymerase chain reaction (PCR) procedure were the large subunit (LSU) and small subunit (SSU) of the ribosomal DNA (rDNA). Primary primers MSP-1 (5´–TGAATGGTCCCTGT–3´) and secondary primers MSP-3 (5´–GGAAT TCACACCGCC CGTCTAT–3´) target the 3 regions of the SSU and recognize a broad range of microsporidian species, including Encephalitozoon spp. and Enterocytozoon bieneusi (8). Primary primers MSP-2B (5´–GTTCA TTCGCACTACT–3´) and secondary primers MSP-4B (5´–CCAAGC TTATGCT TAAGTCCA GGGAG–3´) target the 5 regions of the LSU of E. bieneusi, while primary primers MSP-2A (5´–TCACT CGCCGCTACT–3´) and secondary primers MSP-4A (5´–CCAA GCTTATG CTTAAGTA AGGGT–3´) specifically recognize Encephalitozoon spp. and some other microsporidia but not E. bieneusi. The amplified fragment length by the primers was 300 bp for Encephalitozoon and 500 bp for Enterocytozoon. First, the samples were amplified by the primary primers. Then, the secondary primers were used to differentiate the species of Enterocytozoon and Encephalitozoon. Primary PCRs were performed in a final volume of 25 µL containing 12.5 µL of PCR ready to use Mastermix with 1.5 mM MgCl2 (Bionner), 10 µL of each primer, and 3 µL of template DNA under certain conditions: 95 °C for 5 minutes, followed by 35 cycles of 94 °C for 40 seconds, 55 °C for 45 seconds, and 72 °C for 45 seconds, and a final extension of 72 °C for 4 minutes (8,18).
The second PCRs were performed in a final volume of 25 µL containing 12.5 µL of PCR ready to use Mastermix with 1.5 mM MgCl2 (Bioneer), 10 µL of each primer, and 1 µL of the primary-step PCR product as a template. PCR conditions were 95 °C for 5 minutes, followed by 35 cycles of 94 °C for 40 seconds, 57 °C for E. bieneusi/52 °C for Encephalitozoon spp. for E. bieneusi 35 seconds, 72°C for Encephalitozoon 40 seconds, and 72 °C for 5 minutes as a final extension. Eventually, the restriction fragment length polymorphism (RFLP)-PCR method was used to confirm the existence of Encephalitozoon spp. (8).
Restriction Fragment Length Polymorphism-Polymerase Chain Reaction
RFLP-PCR is utilized to accurately identify the species. It was performed in a final volume of 15 µl containing 6 µL of product primary PCR, 6.5 µL of water, 1.5 µL of buffer, and 1 µL of the Mnl1 restriction enzyme. RFLP-PCR precisely determined Encephalitozoon spp. Encephalitozoon cuniculi will have a distinct restriction pattern (90 and 210 bp), compared to Encephalitozoon intestinalis (60 and 160 bp), while Encephalitozoon hellem will have a distinct pattern consisting of 80 and 180 bp bands (8,19).
Sequencing
Finally, some positive samples were sequenced (Bioneer Company) and compared with the other existing sequences by using the Basic Local Alignment Search Tool and DNAsp, and a phylogenetic tree was drawn using MEGA6 (Neighbor-Joining methods) with 1000 replications as bootstrap support.
Results
From the total samples, 105 (52.5%) cases were females, and 95 (47.5%) of them were males. Out of them, 19 samples were positive, of which 10 samples (5%) were females and 9 samples (4.5%) were males. Of these 19 samples, in 17 samples, the amplification of a band of about 500 bp after PCR indicated the presence of E. bieneusi, and a band of about 300 bp represented the presence of Encephalitozoon spp. in 2 samples, which was further confirmed by enzymatic digestion with Mnl l, resulting in two bands of 90 and 210 bp. and indicating the presence of E. cuniculi. The results of sequence analysis from all positive samples showed the genotype frequencies of D (12) and J (7), respectively (Figures 1 and 2).
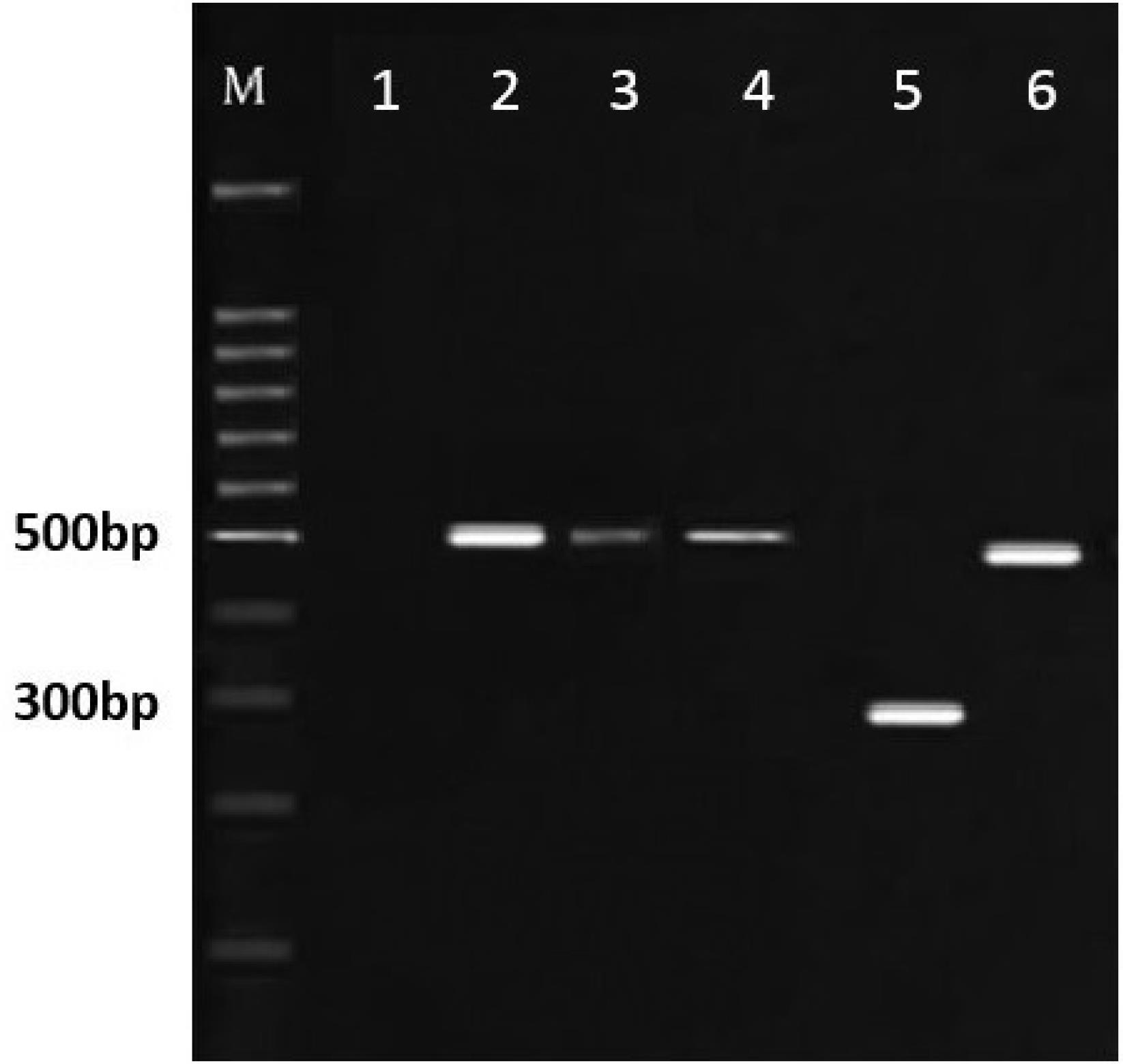
Figure 1.
Electrophoresis of the Target Gene Fragments of Microsporidia. Note. Lane 1: Negative control; Lanes 2-4: Samples of Enterocytozoon bieneusi with bands of 500 bp; Lane 5: Encephalitozoon spp. with band 280 bp; Lane 6: Positive control (DNA of Enterocytozoon bieneusi); M: 100 bp ladder marker
.
Electrophoresis of the Target Gene Fragments of Microsporidia. Note. Lane 1: Negative control; Lanes 2-4: Samples of Enterocytozoon bieneusi with bands of 500 bp; Lane 5: Encephalitozoon spp. with band 280 bp; Lane 6: Positive control (DNA of Enterocytozoon bieneusi); M: 100 bp ladder marker
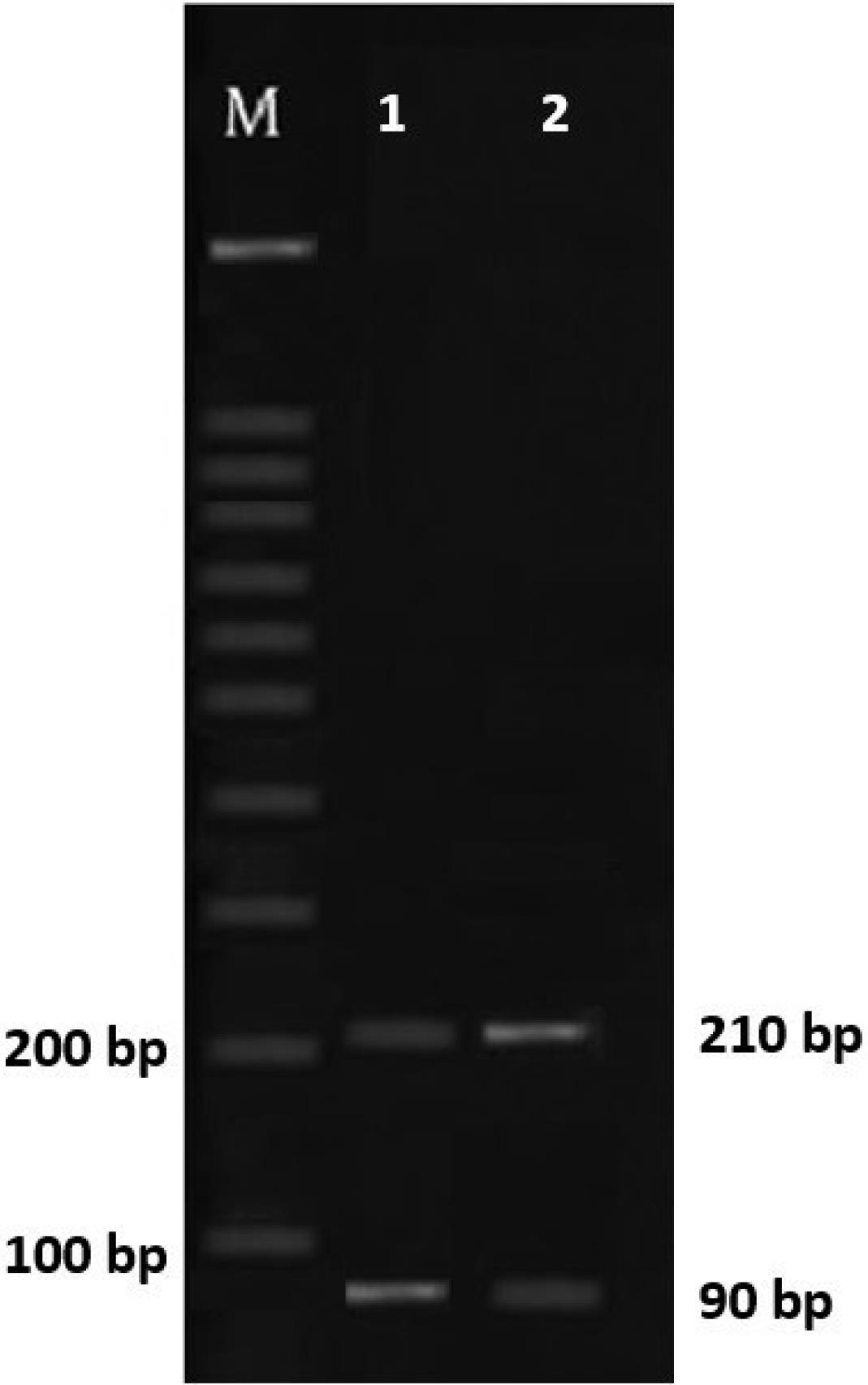
Figure 2.
Electrophoresis of Encephalitozoon spp. Restriction Pattern Using Mnl 1: Lanes 1 and 2: Digest Showing Bands of 210 and 90 bp, Confirming Encephalitozoon cuniculi, and M: 100 bp Ladder Marker
.
Electrophoresis of Encephalitozoon spp. Restriction Pattern Using Mnl 1: Lanes 1 and 2: Digest Showing Bands of 210 and 90 bp, Confirming Encephalitozoon cuniculi, and M: 100 bp Ladder Marker
Phylogenetic Tree
The phylogenetic relationship of Iranian isolates of microsporidia was checked with isolates from other countries accessible from GenBank (Iran isolate: KJ414443, China isolate: KF271490, Malaysia isolates: MH027443, and Austria isolate: DQ793212) by utilizing the neighbor joining method with 1000 replication as bootstrap support (Figure 3).
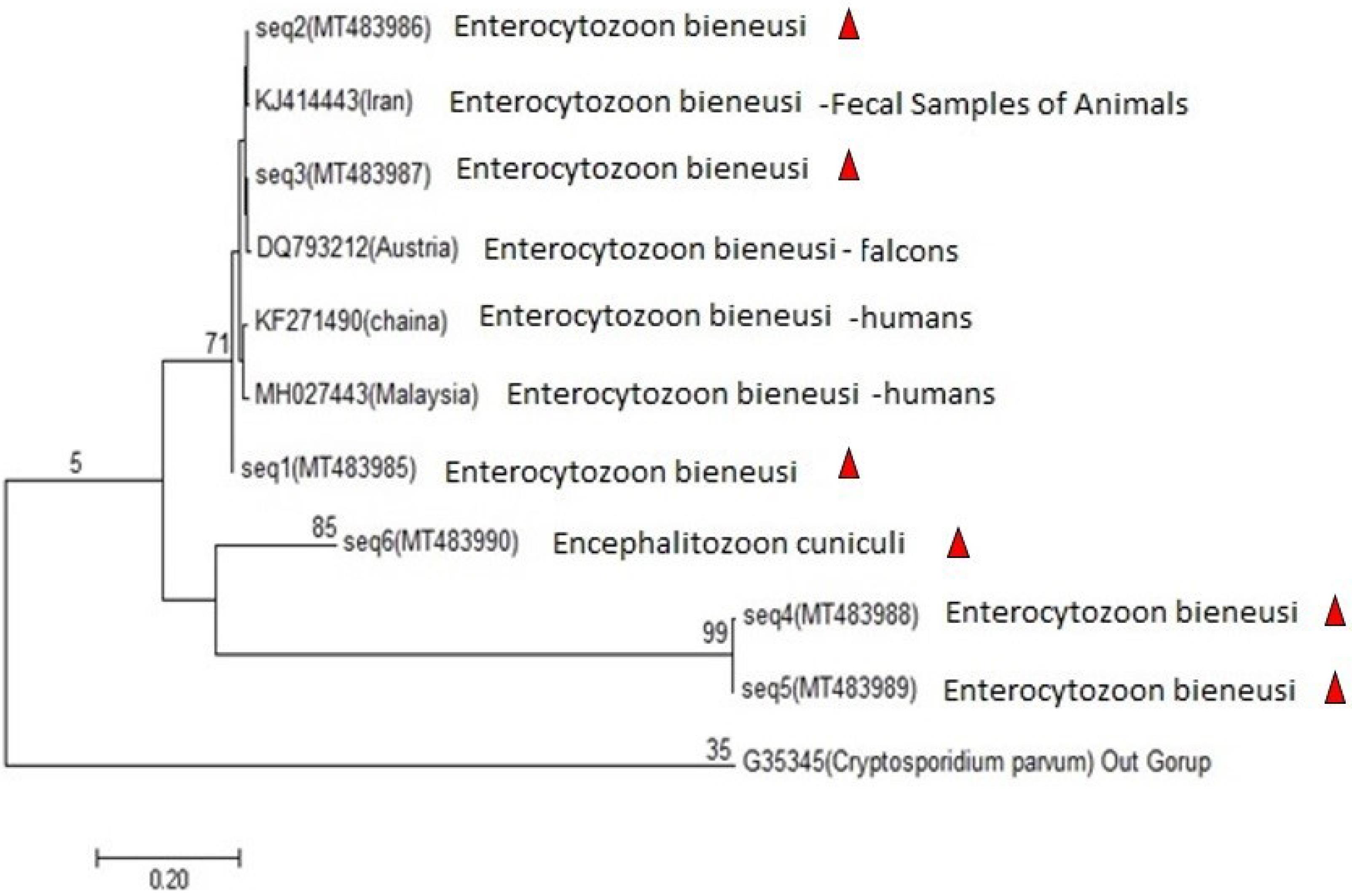
Figure 3.
Phylogenetic Tree of Microsporidia Sequences. Note. A neighbor-joining tree was used for the tree construction of microsporidia from the Iran sequence (1-6) and a few samples from Iran (KJ414443), China (KF271490), Malaysia (MH027443), and Austria (DQ793212). The partial sequence of Cryptosporidium parvum was applied as an outgroup species
.
Phylogenetic Tree of Microsporidia Sequences. Note. A neighbor-joining tree was used for the tree construction of microsporidia from the Iran sequence (1-6) and a few samples from Iran (KJ414443), China (KF271490), Malaysia (MH027443), and Austria (DQ793212). The partial sequence of Cryptosporidium parvum was applied as an outgroup species
Discussion
Microsporidia are an opportunistic zoonotic parasite found all over the world, reported in Iran (11,20). The results of this study demonstrated that, out of 200 samples examined, 19 (9.5%) tested positive for microsporidia by PCR. In this study, 17 samples were related to E. bieneusi, and two samples were Encephalitozoon spp. Then, the Mnl l enzyme was used to identify Encephalitozoon spp. Kord-Sarkachi et al conducted the same study on slaughtered cows in Iran. Both Enterocytozoonspp. and Encephalitozoon spp. were found, and the percentage of Enterocytozoonspp. was higher than that of Encephalitozoon spp. (9), which is consistent with the results of the present study. Askari et al examined the prevalence and genetic diversity of microsporidia in the feces of animals in close contact with humans in Iran. They documented that out of 142 samples, 10.56% were positive (21), which is in line with the results of the current study. Xin-Li Zheng et al performed a study on cattle from the Hainan province of China, analyzing 314 fecal samples from cattle. The prevalence of E. bieneusi was 9.9% (22), which conforms to the findings of the present study. Likewise, Wei Zhao et al evaluated dairy cows in Heilongjiang Province, China and concluded that the rate of infection was 30.1% and only contained E. bieneusi (23). In a descriptive study, Pirestani et al analyzed 126 fecal samples from cows slaughtered in Tehran and reported a 15.1% rate of infection (24), which is higher than the results in the present study. Similarly, Lee et al showed that out of 538 samples of cow feces in South Korea, approximately 81 samples (15%) were positive (25), which is higher than the results of the current study. Nadia Abarca et al analyzed 336 fecal samples from cows in the province of Álava, Northern Spain and found that the prevalence of E. bieneusi was 0.6%. Very few positive samples were reported in their study (26). In another study by Song et al, 490 Holstein cows and 351 dairy buffalo fecal samples were collected in Yunnan province, China. The prevalence of E. bieneusi was 0.59%, and very few positive samples were obtained in this study (27). The phylogenetic analysis revealed that the positive samples identified in this study were divided into three branches. The top branch contained seven samples from Iran, Austria, Chania, and Malaysia and three isolate sequences (1, 2, and 3), and the middle one consisted of three sample sequences (4, 5, and 6), and the lower branch contained an outgroup sample. The possibility of transmission of microsporidiosis increases due to the close relationship between cows and humans. Further, the parasite has the potential to cause disease, making the microsporidiosis of particular importance to human health.
Conclusion
The results revealed that microsporidia were present in cow feces. In addition, these findings indicated that cows can be considered a source of contamination for microsporidia. This disease is zoonosis; Further, it is extremely important to pay attention to the presence of this parasite in domestic animals that are in contact with humans. Further studies should be conducted in different regions and on various animals to understand the epizootiology of the pathogen. Finally, the wide host range of microsporidia requires accurate identification of species and genera in all hosts all over the world.
Authors’ Contribution
Conceptualization: Amin Shafiee, Aminallah Saadatnia, Gholamreza Shokoohi.
Investigation: Ahmad Abolghazi.
Methodology: Aminallah Saadatnia, Gholamreza Shokoohi, Ahmad Abolghazi.
Writing–original draft: Amin Shafiee, Aminallah Saadatnia, Gholamreza Shokoohi, Ahmad Abolghazi.
Writing–review & editing: Amin Shafiee, Aminallah Saadatnia, Gholamreza Shokoohi, Ahmad Abolghazi.
Competing Interests
The authors declare no conflict of interests.
Consent for Publication
Not applicable.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
This study was approved by the Ethics Committee of Jahrom University of Medical Sciences (with code: IR.JUMS.REC.1397.141).
Funding
The study was financially supported by a grant from Jahrom University of Medical Sciences (grant no. 1397.141).
References
- Snowden KF, Shadduck JA. Microsporidia in higher vertebrates. In: Wittner M, Weiss LM, eds. The Microsporidia and Microsporidiosis. Washington, DC: ASM Press; 1999. p. 393-417.
- Pan G, Bao J, Ma Z, Song Y, Han B, Ran M. Invertebrate host responses to microsporidia infections. Dev Comp Immunol 2018; 83:104-13. doi: 10.1016/j.dci.2018.02.004 [Crossref] [ Google Scholar]
- Weiser J. Microsporidia in invertebrates: host-parasite relations at the organismal level. In: Bulla LA, Cheng TC, eds. Biology of the Microsporidia. Boston, MA: Springer; 1976. p. 163-201. 10.1007/978-1-4684-3114-8_7.
- Sprague V, Becnel JJ, Hazard EI. Taxonomy of phylum microspora. Crit Rev Microbiol 1992; 18(5-6):285-395. doi: 10.3109/10408419209113519 [Crossref] [ Google Scholar]
- Thellier M, Breton J. Enterocytozoonbieneusi in human and animals, focus on laboratory identification and molecular epidemiology. Parasite 2008; 15(3):349-58. doi: 10.1051/parasite/2008153349 [Crossref] [ Google Scholar]
- Mathis A, Weber R, Deplazes P. Zoonotic potential of the microsporidia. Clin Microbiol Rev 2005; 18(3):423-45. doi: 10.1128/cmr.18.3.423-445.2005 [Crossref] [ Google Scholar]
- Franzen C, Müller A. Microsporidiosis: human diseases and diagnosis. Microbes Infect 2001; 3(5):389-400. doi: 10.1016/s1286-4579(01)01395-8 [Crossref] [ Google Scholar]
- Conteas CN, Berlin OG, Ash LR, Pruthi JS. Therapy for human gastrointestinal microsporidiosis. Am J Trop Med Hyg 2000; 63(3-4):121-7. doi: 10.4269/ajtmh.2000.63.121 [Crossref] [ Google Scholar]
- Kord-Sarkachi E, Tavalla M, Beiromvand M. Molecular diagnosis of microsporidia strains in slaughtered cows of southwest of Iran. J Parasit Dis 2018; 42(1):81-6. doi: 10.1007/s12639-017-0969-4 [Crossref] [ Google Scholar]
- Karimi K, Mirjalali H, Niyyati M, Haghighi A, Pourhoseingholi MA, Sharifdini M. Molecular epidemiology of Enterocytozoonbieneusiand Encephalitozoon sp, among immunocompromised and immunocompetent subjects in Iran. Microb Pathog 2020; 141:103988. doi: 10.1016/j.micpath.2020.103988 [Crossref] [ Google Scholar]
- Ghoyounchi R, Ahmadpour E, Spotin A, Mahami-Oskouei M, Rezamand A, Aminisani N. Microsporidiosis in Iran: a systematic review and meta-analysis. Asian Pac J Trop Med 2017; 10(4):341-50. doi: 10.1016/j.apjtm.2017.03.017 [Crossref] [ Google Scholar]
- Didier ES, Weiss LM. Microsporidiosis: current status. Curr Opin Infect Dis 2006; 19(5):485-92. doi: 10.1097/01.qco.0000244055.46382.23 [Crossref] [ Google Scholar]
- Wasson K, Peper RL. Mammalian microsporidiosis. Vet Pathol 2000; 37(2):113-28. doi: 10.1354/vp.37-2-113 [Crossref] [ Google Scholar]
- Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop 2005; 94(1):61-76. doi: 10.1016/j.actatropica.2005.01.010 [Crossref] [ Google Scholar]
- Wittner M, Weiss LM. The Microsporidia and Microsporidiosis. Washington, DC: ASM Press; 1999.
- Fars Province Planning and Budget Organization. Deputy of Statistics and Information. Inst Med; 2016. p. 156-62.
- Fedorko DP, Hijazi YM. Application of molecular techniques to the diagnosis of microsporidial infection. Emerg Infect Dis 1996; 2(3):183-91. doi: 10.3201/eid0203.960304 [Crossref] [ Google Scholar]
- Velásquez JN, Carnevale S, Guarnera EA, Labbé JH, Chertcoff A, Cabrera MG. Detection of the microsporidian parasite Enterocytozoonbieneusi in specimens from patients with AIDS by PCR. J Clin Microbiol 1996; 34(12):3230-2. doi: 10.1128/jcm.34.12.3230-3232.1996 [Crossref] [ Google Scholar]
- Sironi M, Bandi C, Novati S, Scaglia M. A PCR-RFLP method for the detection and species identification of human microsporidia. Parassitologia 1997; 39(4):437-9. [ Google Scholar]
- Stentiford GD, Becnel J, Weiss LM, Keeling PJ, Didier ES, Williams BP. Microsporidia - emergent pathogens in the global food chain. Trends Parasitol 2016; 32(4):336-48. doi: 10.1016/j.pt.2015.12.004 [Crossref] [ Google Scholar]
- Askari Z, Mirjalali H, Mohebali M, Zarei Z, Shojaei S, Rezaeian T. Molecular detection and identification of zoonotic microsporidia spore in fecal samples of some animals with close-contact to human. Iran J Parasitol 2015; 10(3):381-8. [ Google Scholar]
- Zheng XL, Zhou HH, Ren G, Ma TM, Cao ZX, Wei LM. Genotyping and zoonotic potential of Enterocytozoonbieneusi in cattle farmed in Hainan province, the southernmost region of China. Parasite 2020; 27:65. doi: 10.1051/parasite/2020065 [Crossref] [ Google Scholar]
- Zhao W, Zhang W, Yang F, Zhang L, Wang R, Cao J. Enterocytozoonbieneusi in dairy cattle in the Northeast of China: genetic diversity of ITS gene and evaluation of zoonotic transmission potential. J Eukaryot Microbiol 2015; 62(4):553-60. doi: 10.1111/jeu.12210 [Crossref] [ Google Scholar]
- Pirestani M, Sadraei J, Forouzandeh M. Molecular characterization of zoonotic isolates of Enterocytozoonbieneusi in Iran. Feyz 2012;16(1):51-7. [Persian].
- Lee JH. Prevalence and molecular characteristics of Enterocytozoonbieneusi in cattle in Korea. Parasitol Res 2007; 101(2):391-6. doi: 10.1007/s00436-007-0468-0 [Crossref] [ Google Scholar]
- Abarca N, Santín M, Ortega S, Maloney JG, George NS, Molokin A. Molecular detection and characterization of Blastocystis sp and Enterocytozoonbieneusi in cattle in Northern Spain. Vet Sci 2021; 8(9):191. doi: 10.3390/vetsci8090191 [Crossref] [ Google Scholar]
- Song HY, Wang KS, Yang JF, Mao HM, Pu LH, Zou Y. Prevalence and novel genotypes identification of Enterocytozoonbieneusi in dairy cattle in Yunnan province, China. Animals (Basel) 2021; 11(11):3014. doi: 10.3390/ani11113014 [Crossref] [ Google Scholar]