Avicenna Journal of Clinical Microbiology and Infection. 10(3):112-119.
doi: 10.34172/ajcmi.3458
Original Article
The Molecular Investigation of the mecA Gene and Antibiotic Susceptibility Pattern of Staphylococcus aureus and Staphylococcus epidermidis Isolated from Patients with Immune System Disorders at Omid Hospital, Isfahan, Iran
Zahra Babaei 1  , Monir Doudi 2, *
, Monir Doudi 2, *  , Ladan Rahimzadeh Torabi 2
, Ladan Rahimzadeh Torabi 2 
Author information:
1Department of Microbiology, Naein Branch, Islamic Azad University, Isfahan, Iran
2Department of Microbiology, Falavarjan Branch, Islamic Azad University, Isfahan, Iran
Abstract
Background: At present, antibiotic-resistant staphylococci, especially methicillin-resistant strains, are prevalent agents of infections in medical centers and hospitals. The objective of the present investigation was to discern and trace the methicillin resistance gene harbored in two bacterial strains, namely Staphylococcus aureus and Staphylococcus epidermidis, obtained from clinical specimens gathered from patients exhibiting immune system deficiency at Omid hospital located in Isfahan.
Methods: The present investigation was conducted utilizing a descriptive cross-sectional approach. Initially, a total of 70 clinical isolates comprising 35 isolates of S. aureus and 35 isolates of S. epidermidis were obtained from patients who were diagnosed with immunodeficiency and admitted to Omid Hospital located in Isfahan, Iran, from January 2017 to April 2018. After the characterization of the isolates via morphological and biochemical assessments, subsequent evaluation of their antibiotic sensitivity was performed through the utilization of disk diffusion and Epsilometer test (E-test). Then, the identification of the isolates was conducted using the colony PCR method incorporating primers (MCF, MCR, GAIF, and GAIR) and elucidated through molecular analysis.
Results: In this study, all isolates of S. aureus were resistant to cefoxitin and the MIC of this antibiotic was confirmed using E-test. However, of 35 S. epidermidis isolates, 30 isolates (85.7%) were resistant to oxacillin and 5 isolates (14.3%) were sensitive to oxacillin. According to the molecular findings, out of 35 isolates of methicillin-resistant S. aureus, 4 isolates (11.4%) had the mecA gene, and out of 35 isolates of S. epidermidis, 10 isolates (28.5%) had the mecA gene.
Conclusion: The present study revealed that precise detection of methicillin resistance in the aforementioned bacterial strains necessitates the employment of both phenotypic and genotypic methods. The frequency of the mecA gene in methicillin-resistant S. aureus (MRSA) was found to be declining. The incidence of methicillin-resistant S. epidermidis (MRSE) is on the rise.
Keywords: Methicillin-resistant Staphylococcus aureus, Methicillin-resistant Staphylococcus epidermidis, mecA gene, PCR, E-test, Disc diffusion
Copyright and License Information
© 2023 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Babaei Z, Doudi M, Rahimzadeh Torabi L. The molecular investigation of the mecA gene and antibiotic susceptibility pattern of Staphylococcus aureus and Staphylococcus epidermidis isolated from patients with immune system disorders at Omid Hospital, Isfahan, Iran. Avicenna J Clin Microbiol Infect. 2023; 10(3):112-119. doi:10.34172/ajcmi.3458
Introduction
Staphylococcus aureus and Staphylococcus epidermidis are widely recognized as ubiquitous and pathogenic bacteria implicated in hospital-acquired infections on a global scale (1-3). S. aureus has been observed to colonize on the skin, particularly when damaged, as well as in various other areas including the perineum, vagina, armpit, navel of neonates, and oropharynx (4,5). This gram-positive bacterium represents a significant causative agent in the domain of healthcare-associated infections (6). Methicillin-resistant S. aureus (MRSA) is a strain of S. aureus that is resistant to methicillin and other beta-lactam antibiotics. Methicillin-resistant S. aureus (MRSA) constitutes a grave concern in the context of hospital-acquired infections, owing to its opportunistic nature, and exacerbates the complexity of treating infections caused by this bacterium (7,8). One of the contributory factors for the resistance of the bacteria to methicillin is the indiscriminate administration of antibiotics, particularly beta-lactam antibiotics. This may be attributed to the inadequate and uninformed prescription practices of physicians or the non-performance of an antibiogram (9,10). The prevalence of methicillin resistance among S. epidermidis isolated from hospital samples ranges from 75% to 90% (11). Usually, the carriers are the reservoir of antibiotic resistance genes and cause commensal S. epidermidis to become a pathogen. Most antibiotic resistance genes are coded by plasmids and can be transferred from methicillin-resistant strains to sensitive strains (12). Generally, these plasmids carry antibiotic resistance genes. According to findings obtained from the investigation conducted by researchers, it was revealed that the occurrence of MRSA strains was reported in European medical facilities from 1961 to 1963, a few years after their introduction (13,14). The prevalence rates of this phenomenon have been reported to exceed 70% in Asian nations, including China, Korea, and Taiwan while surpassing 50% in North America and Iran and attaining 20% in Europe (15). Methicillin is a semi-synthetic penicillinase-resistant penicillin. Methicillin resistance is one of the most important and common resistance patterns among S. aureus strains that is caused by the presence of the mecA gene, which is chromosomally coded (16,17). The development of resistance to antimicrobial agents in MRSA strains has been attributed to the production of a distinctive binding protein referred to as Penicillin Binding Protein 2a (PBP2a). This protein has a substantially weakened affinity towards β-lactam antibiotics, thus prompting the development of bacterial strains that are more resistant to these therapeutic agents (18-20). PBP2a is encoded by the mecA gene which is located on a large mobile genetic element called staphylococcal cassette chromosome mec (SCCmec) and is present in the chromosome of resistant strains (21-24). According to the reports of the World Health Organization, patients who are infected with MRSA are hospitalized for a longer period of time than those who are infected with methicillin sensitive S. aureus (MSSA). Therefore, in addition to the cost of treatment, the infection can progress to bacteremia or endocarditis (25,26). Complications of infection such as kidney and liver failure are also more prevalent among MRSA patients than among patients infected with MSSA (27,28) and it has even been observed that the mortality rate is significantly higher among patients infected with MRSA, especially those with immune system defects, cancer patients, transplant recipients, AIDS patients, elderly, infants, pregnant women, diabetics, and so on, than among MSSA patients (29-33). Timely diagnosis and isolation of these patients can prevent the spread of MRSA and MSSA strains in the hospital environment and medical staff. Based on statistical analysis, it has been determined that over 70% of S. epidermidis strains isolated from hospitals have been found to exhibit resistance to methicillin. Furthermore, the majority of these strains have demonstrated a level of multidrug resistance, which makes their treatment difficult and expensive (34-36). The objective of this study was to analyze the mecA gene and antibiotic susceptibility pattern in S. aureus and S. epidermidis strains isolated from patients with immune system disorders at Omid Hospital in Isfahan, Iran. The disk diffusion and E-test methods were employed to isolate the gene, followed by the colony PCR method for identification of the mecA gene.
Materials and Methods
Clinical Isolates
A total of 70 isolates of MRSA and MRSE isolated from clinical samples of patients with immune system deficiency in Omid hospital in Isfahan were evaluated. These isolates were from different clinical samples, including wounds, sputum, urine, blood, trachea, and so on. Immunodeficiency patients (cancer patients, transplant patients, AIDS patients, the elderly, infants, pregnant women, diabetic patients, etc) were collected and standard tests were performed to isolate and identify methicillin-resistant isolates, which included preparation of Gram staining slides, culture in mannitol salt agar, DNAse agar and coagulase and catalase tests as well as antibiogram against cefoxitin, oxacillin, vancomycin, novobiocin, bacitracin antibiotics, and cefoxitin, oxacillin, and vancomycin E-tests (37).
Disc Diffusion Method
The disk release test was performed using disks (Mast, England) containing cefoxitin (30 μg), bacitracin (0.04 μg), vancomycin (30 μg), and novobiocin (5 μg). Two strains of S. aureus (ATCC 33591 and ATCC 25923) and two strains of S. epidermidis (ATCC 29887 and ATCC 12228) were used for positive and negative control. The antibiotic sensitivity test was performed using the Kirby-Bayer disk diffusion method on Mueller Hinton agar (MHA) medium, according to the Clinical and Laboratory Standards Institute (CLSI ) guidelines (38).
Determination of MIC Using E-test Method
The E-test was performed on each isolate individually to verify the results of the disk diffusion method. For this test, a bacterial suspension equal to 0.5 McFarland standard was prepared and cultured on MHA medium using sterile swap in four directions. Then, each E-test strip was placed separately on the plate. In this study, E-test belonging to (Mast, England), cefoxitin, vancomycin, and oxacillin were used for the isolates. Two strains of S. aureus (ATCC 33591 and ATCC 25923) and two strains S. epidermidis (ATCC 29887 and ATCC 12228) were used again for positive and negative control (37,39).
Colony-PCR and Identification of its Products
Colony PCR is a method of DNA fragment amplification by PCR, which is done using a microorganism colony without the need for DNA extraction. The first step of unwinding DNA double helix into two single strands of DNA for the first primer is done at 94 °C for 300 seconds, followed by 40 cycles of amplification, including the denaturation step at 94 °C for 15 seconds, the annealing step at 55 °C for 15 seconds, and the extension step at 72 °C for 20 seconds, and the final elongation step at 72 °C for 300 seconds. The first step of unwinding DNA double helix into two single-stranded DNA molecules for the second primer is done at 94 °C for 300 seconds, followed by 40 amplification cycles, including the denaturation step at 94 °C for 30 seconds, the annealing step at 55 °C for 30 seconds, and the extension step at 72 °C for 60 seconds, and the final elongation step at 72 °C for 300 seconds. It should be noted that the positive control had the mecA gene and the negative control did not have this gene. Moreover, sterile distilled water can be used for negative control (40,41). The primer sequences used in the present investigation are delineated in Table 1.
Table 1.
The Primers Employed in this Study for Detecting the mecA Gene
|
Primers
|
(3´) Primer Sequence (5´)
|
Product (bp)
|
|
GaiF
|
AAAATCGATGGTAAAGGTTGGC |
307 |
|
GaiR
|
AGTTCTGCAGTACCGGATTTGC |
|
|
MCF
|
TGGCTATCGTGTCACAATCG |
500 |
|
MCR
|
CTGGAACTTGTTGAGCAGAG |
|
Statistical Analysis
In order to statistically analyze the data obtained from this research, Chi-square test was used in SPSS (version 17) at the confidence level of P ≤ 0.05.
Results
In general, in this study, out of 70 isolates of Staphylococcus, including 35 isolates of S. aureus and 35 isolates of S. epidermidis, were resistant to methicillin. They were isolated from surgery, ICU, operating room, neurology, and gastroenterology departments of Omid hospital in Isfahan. Out of 35 S. aureus isolates, 21 samples belonged to women (60%) and 14 samples belonged to men (40%). Of 35 isolates of S. epidermidis, 20 samples belonged to women (57.2%) and 15 samples belonged to men (42.8%). The clinical samples were collected from patients with immune system deficiency (diabetics, AIDS patients, transplant recipients, elderly, newborns, pregnant women, etc.). The highest frequency of S. aureus strain was observed in the trachea of 17 samples (48.5%) and the lowest frequency was observed in infectious eye discharge (1 sample, 2.8%) and urine (1 sample, 2.8%). The highest frequency of S. epidermidis strain was observed in the blood of 24 samples (68.5%), and the lowest frequency was observed in the catheter of 1 sample (2.8%) and the cerebrospinal fluid of 1 sample (2.8%)
The Results of Identification of Isolates Using Phenotypic and Biochemical Tests
In this study, the identified isolates, including S. epidermidis and S. aureus, were observed as gram-positive cocci and as single or double or irregular grape-shaped clusters using gram staining. After identifying the morphology and arrangement of bacteria with the help of gram staining, the identification of these bacteria was achieved through the application of targeted biochemical assays.
Results of the Antibiotic Sensitivity Test by Disk Diffusion Method
The results of the antibiotic sensitivity test of clinical isolates of S. aureus and S. epidermidis are presented in Table 2 and Figure 1. In this research, out of a total of 35 clinical isolates of S. aureus, all 35 isolates (100%) were resistant to both cefoxitin and bacitracin, and 100% of the isolates were sensitive to vancomycin and novobiocin. Out of a total of 35 clinical isolates of S. epidermidis, 4 isolates (11.4%) were sensitive to cefoxitin and 31 isolates (88.5%) were resistant to cefoxitin. All 35 isolates (100%) were resistant to bacitracin and 10 isolates (28.5%) were sensitive to vancomycin. Additionally, 25 isolates (71.4%) were resistant to vancomycin and all 35 isolates (100%) were sensitive to novobiocin.
Table 2.
Antibiotic Sensitivity and Resistance Pattern of MRSA and MRSE Isolates According to the Type of Antibiotic by Disk Diffusion Method
|
Antibiotics
|
Concentration
(µg/mL)
|
MRSA (n=35)
|
MRSE (n=35)
|
Resistance
No. (%)
|
Sensitive
No. (%)
|
Resistance
No. (%)
|
Sensitive
No. (%)
|
| Cefoxitin |
30 |
35 (100) |
- |
31 (88.5) |
4 (11.4) |
| Bacitracin |
0.04 |
35 (100) |
- |
35 (100) |
- |
| Vancomycin |
30 |
27 (77.1) |
8 (22.9) |
25 (71.4) |
10 (28.5) |
| Novobiocin |
5 |
- |
35 (100) |
- |
35 (100) |
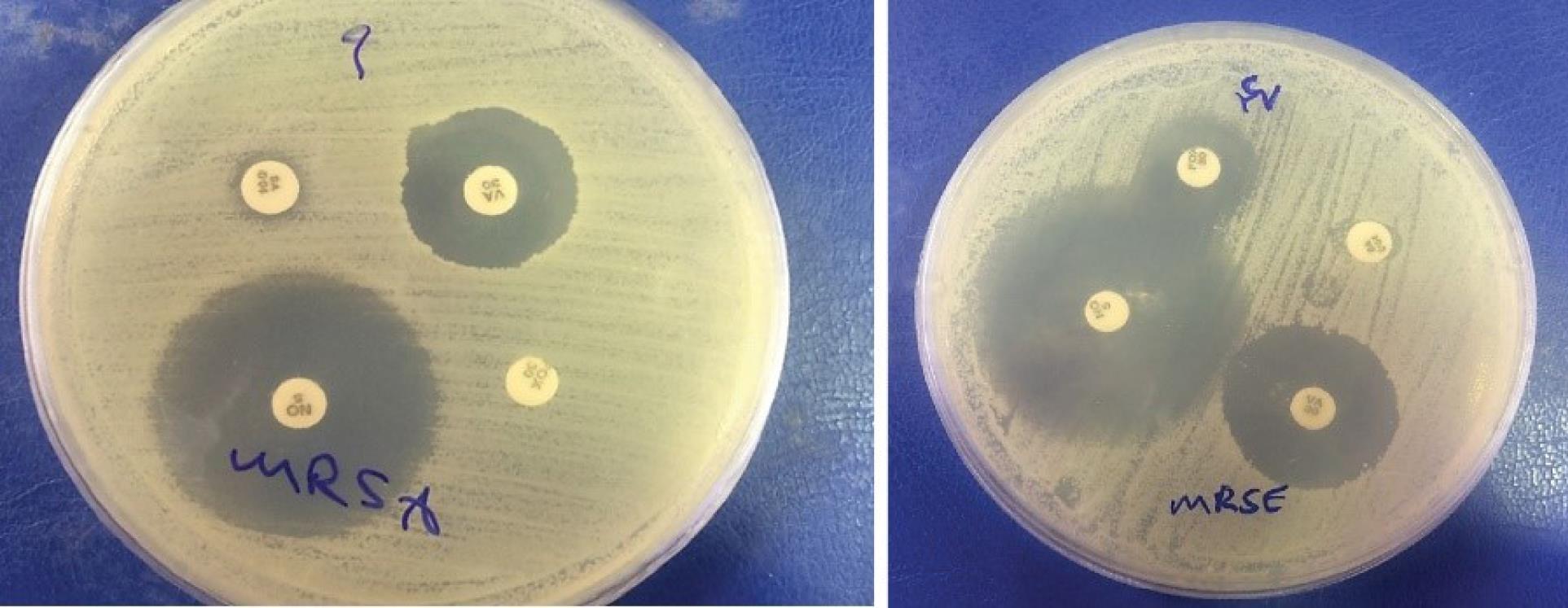
Figure 1.
Antibiogram Result of One Isolate of MRSA and MRSE
.
Antibiogram Result of One Isolate of MRSA and MRSE
E-test Results
The results of this test showed that out of 35 methicillin-resistant isolates, 27 samples (77.1%) were resistant to vancomycin and 8 isolates (22.9%) were sensitive to vancomycin. All strains of MRSA exhibited complete resistance (100%) to cefoxitin as depicted by the results obtained from the E-test strips (Figure 2). The results obtained for S. epidermidis isolates in this study showed that out of 35 methicillin-resistant isolates, 20 samples (57.2%) were resistant to cefoxitin and 15 isolates (42.8%) were sensitive to cefoxitin. Out of 35 isolates of S. epidermidis that exhibited resistance to methicillin, 10 isolates (28.5%) were resistant to vancomycin and 25 isolates (71.5%) were sensitive to vancomycin. Out of 35 isolates of S. epidermidis that exhibited resistance to methicillin, 30 isolates (85.7%) were resistant to oxacillin and 5 samples (14.3%) were determined to be oxacillin-sensitive (Figure 3).
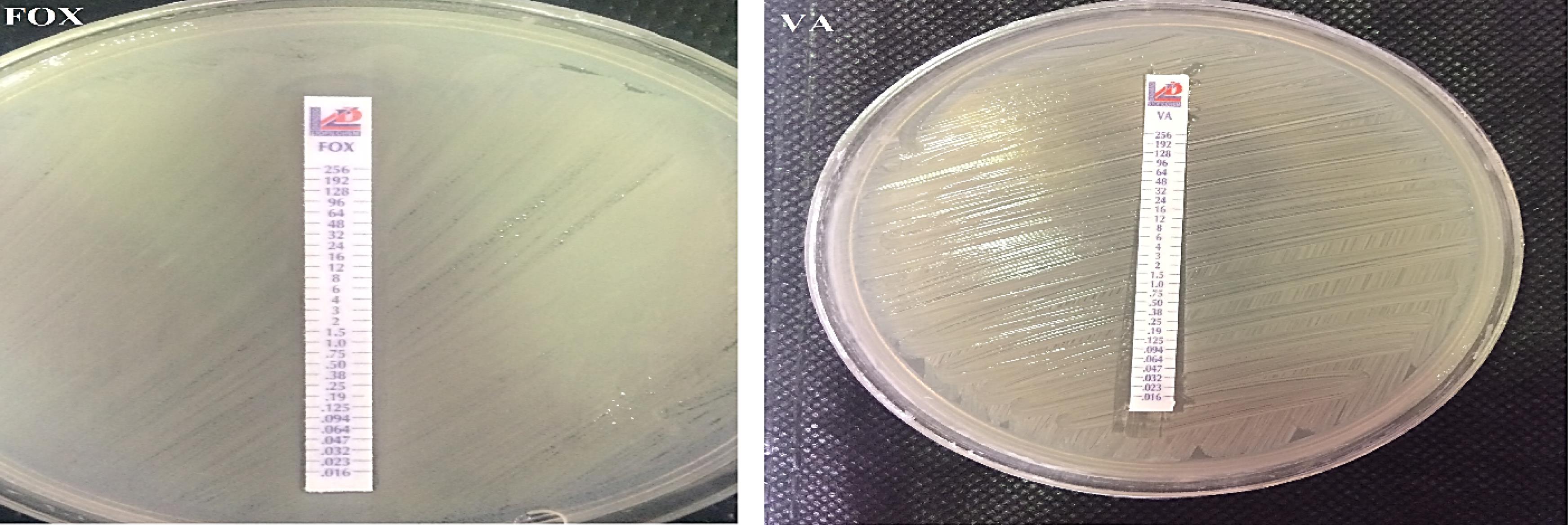
Figure 2.
Determination of MIC for Cefoxitin and Vancomycin Antibiotics in Two MRSA Isolates Using E-test
.
Determination of MIC for Cefoxitin and Vancomycin Antibiotics in Two MRSA Isolates Using E-test
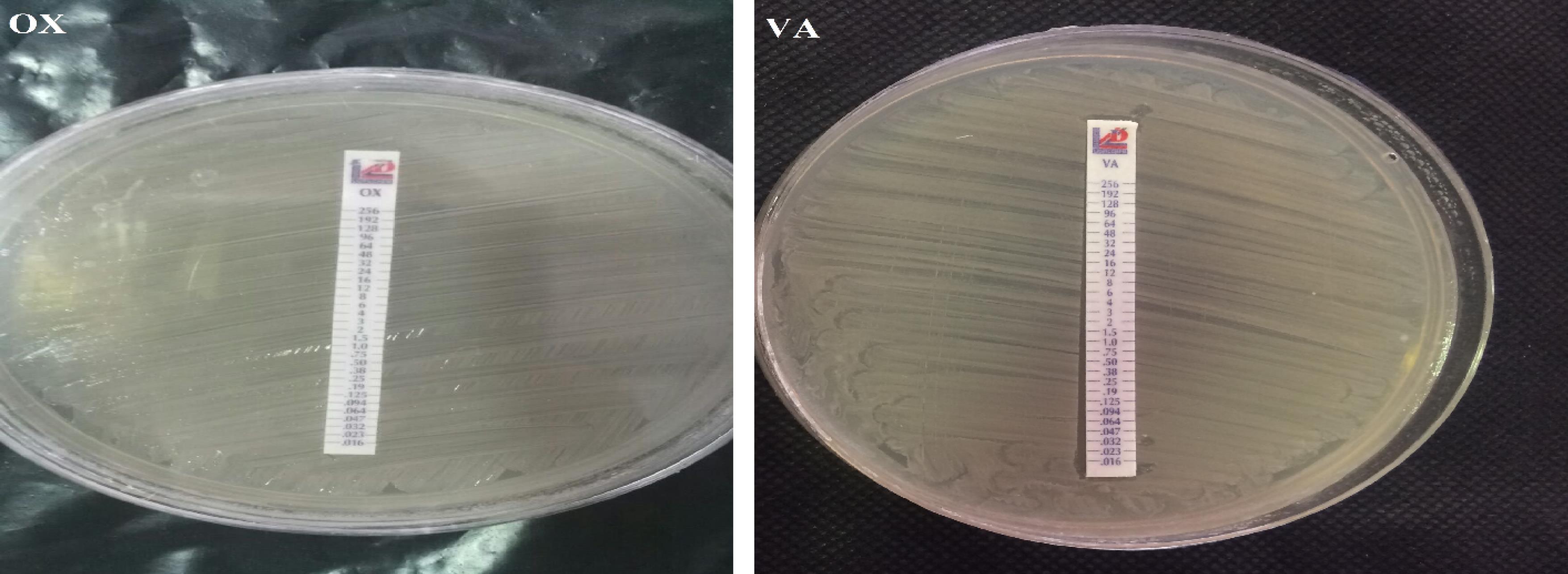
Figure 3.
Determination of MIC for Oxacillin and Vancomycin Antibiotics in Two MRSE Isolates Using E-test
.
Determination of MIC for Oxacillin and Vancomycin Antibiotics in Two MRSE Isolates Using E-test
The Results of Colony PCR to Detect the mecA Gene in MRSAs
In this study, two primers were used for the identification of the mecA gene. The first primer used was only able to identify a part of the mecA gene that was the same in all the variants, and the second primer completed the identification of the mecAgene in the desired variants. After the isolation of methicillin-resistant strains by phenotypic method, the mecA gene was detected by PCR. Two strains of S. aureus (ATCC 33591 and ATCC 25923) were used for positive and negative control, respectively. The obtained results indicated that out of 35 isolates of MRSA, 4 isolates (11.4%) had the mecA gene and 31 isolates (88.6%) lacked the mecAgene (Figure 4).
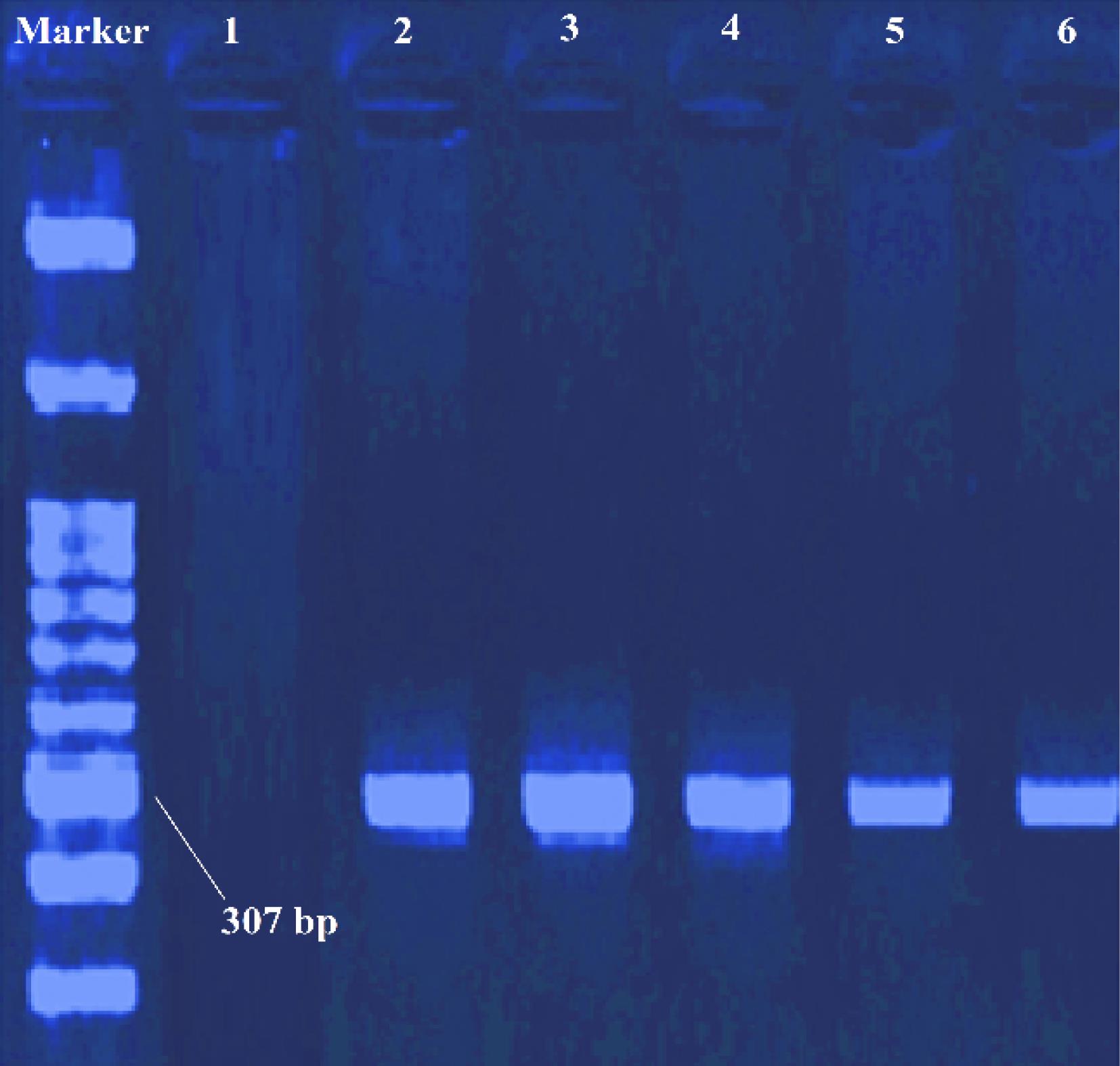
Figure 4.
Results of PCR and the Presence of mecA Gene for Several MRSA Isolates. Note: Lane 2: Weighted marker 100 bp, Lanes 3-5: Clinical samples, Lane 6: Positive control, Lane 1: Negative control
.
Results of PCR and the Presence of mecA Gene for Several MRSA Isolates. Note: Lane 2: Weighted marker 100 bp, Lanes 3-5: Clinical samples, Lane 6: Positive control, Lane 1: Negative control
Results of Colony PCR to Trace the mecA Gene in MRSEs
After the isolation of methicillin-resistant strains by phenotypic method, the mecAgene was detected by PCR. Two strains of S. epidermidis ATCC 29887 and S. epidermidis ATCC 12228 were used for positive and negative control, respectively. The results obtained from 35 isolates of MRSE showed that 10 isolates (28.5%) had the mecA gene and 25 isolates (71.4%) did not have the mecA gene (Figure 5).
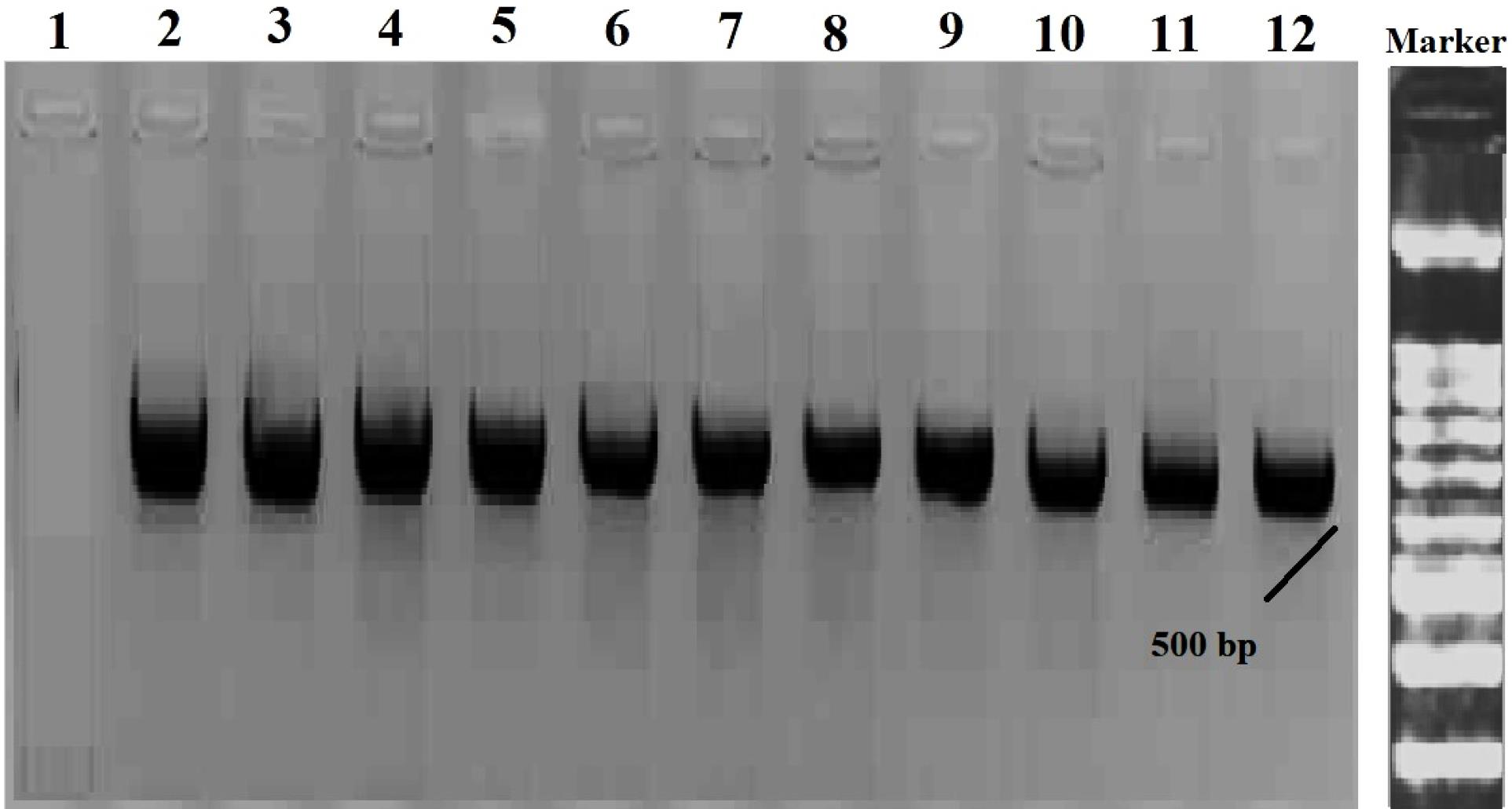
Figure 5.
Results of PCR and the Presence of mecA Gene for Several MRSE Isolates. Note: Well 2: Weighted marker 100 bp, Wells 3-11: Clinical samples, Well 12: Positive control, Well 1: Negative control
.
Results of PCR and the Presence of mecA Gene for Several MRSE Isolates. Note: Well 2: Weighted marker 100 bp, Wells 3-11: Clinical samples, Well 12: Positive control, Well 1: Negative control
Discussion
In recent years, S. aureus and S. epidermidis have been identified as prominent etiological agents of hospital-acquired infections in vulnerable individuals with compromised immune function (42,43). According to a global survey, an observed correlation ranging from 65% to 85% linked clinical strains to MRSE. The increasing incidence of nosocomial infections and the mounting issue of antimicrobial resistance necessitate the prompt implementation of a more expeditious diagnostic method in healthcare facilities. It is recommended that genotypic tests should be replaced with phenotypic tests (44,45). In this research, 35 isolates of S. aureus and 35 isolates of S. epidermidis were selected from 70 clinical isolates in a period of six months. The prevalence of S. aureus strain exhibited significant variability among different sample types, with the trachea demonstrating the highest frequency (48.5%). The S. epidermidis strain with the greatest frequency was observed in the blood of 24 samples, accounting for 68.5% of the total. The current research found that cefoxitin resistance was displayed by MRSA strains, which was later confirmed through an E-test performed to measure the MIC of cefoxitin. However, it should be noted that the E-test of cefoxitin was not explicitly outlined in the CLSI guidelines for MRSE isolates. One of the important reasons that has attracted attention to S. aureus bacteria today is the resistance mechanisms of these bacteria given that the prevalence of MRSA strains in hospitals and medical centers is increasing (46). Pishva et al reported that the rate of resistance to methicillin in S. epidermidis isolates using the agar dilution method was 10.9% and the rate of resistance to oxacillin was 13.5% using the E-test (47). Another study performed by Sharma et al indicated that all the isolates were susceptible to vancomycin, linezolid, and teicoplanin in the disc diffusion test while maximum resistance was noted against penicillin (100%) and 25% of the isolates were found to be resistant to methicillin. A comparison between resistance patterns of methicillin-resistant and methicillin-sensitive strains showed that methicillin-resistant isolates had higher levels of resistance to other antibiotics (48). Based on the results obtained in our study for MRSE isolates, 28.5% of the isolates were sensitive to vancomycin using the E-test, indicating that the sensitivity to this antibiotic was reduced compared to the results of Sharma and Hejira. The results obtained in our study also showed that the analysis of MRSE strains using the E-test method determined 85.7% resistance to oxacillin, indicating a significant increase compared to the results of the study conducted by Pishva et al. The increase in oxacillin resistance is likely a result of individuals intentionally taking antibiotics, particularly antibiotics from the penicillin family such as amoxicillin, ampicillin, nafcillin, and similar ones. In a study conducted by Pishva et al in Isfahan, the resistance rate of S. epidermidis isolates to methicillin in Al-Zahra Hospital was reported to be 73% (47). In other studies, the prevalence of MRSA was determined to be 6.3%. Moreover, among all isolates of S. aureus, the prevalence of MRSA was observed to be 61.8%. The presence of mec genes was observed in 96.8% of MRSA isolates, with the remaining 3.2% exhibiting an absence of mec genes. The co-occurrence of mecA and mecC was identified in 57.1% of the MRSA isolates. The antibiotics that exhibited the highest level of resistance were penicillin and amoxicillin/clavulanic acid, followed by norfloxacin, levofloxacin, ciprofloxacin, azithromycin, erythromycin, moxifloxacin, and sulfamethoxazole/trimethoprim, with resistance rates of 91.2%, 87.1%, 83.9%, 78.6%, 77.4%, 69.8%, and 54.9%, respectively. In contrast, vancomycin and teicoplanin displayed high efficacy, with a success rate of 98.4% in combating MRSA (49). Our findings showed that out of 35 MRSA isolates, only 4 isolates (11.4%) had the mecA gene and out of 35 MRSE isolates, 10 isolates (28.5%) had the mecA gene, indicating a lower frequency of this gene in MRSE isolates compared to the studies conducted between 2016 and 2018 (48-52) but a higher frequency compared to the results of the study conducted by Rahimi in 2012 (53). Kondo et al isolated 99 strains of S. aureus from America, Canada, England, Ireland, and Europe and after performing microbial and PCR testing, the findings revealed that 16 of the isolates were found to have the mecA gene (54). In another study, researchers sampled 50 patients who underwent joint plastic surgery. These patients were examined after receiving antibiotics for a period of 24, 36, and 48 months, and their clinical samples were taken. It was determined that 38 patients had MRSE and 12 patients had MRSA (55). Prasad et al studied patients with implant-related infections. A total of 91 clinical samples were obtained and biochemical and PCR tests were performed. Of 55 (42.2%) isolates of S. epidermidis, 23 (41.8%) samples were multiple drug-resistant and 15.3% of them were resistant to methicillin and had the mecA gene (56). In another study, 26 nasal samples were collected from premature babies and antibiotic and microbial tests along with PCR were performed on the isolates. Based on the results of the PCR test, the most frequently isolated species were S. epidermidis (38.3%) and S. haemolyticus (38%), followed by other Staphylococcus species. It should be noted that the isolated staphylococci had multiple drug resistance and some of them had the mecA gene (57). In the study conducted by Du et al, a variety of microbial strains were isolated from hospitalized patients, outpatients, and hospital personnel. Subsequently, the isolates were subject to biochemical and antibiotic analyses employing two distinct methods, namely E-test and disk diffusion, along with molecular identification techniques. According to the results obtained from their research, 44.8% of isolates were MRSE and MRSE-ST2-SCCmecIII was the predominant clone in clinical isolates, almost resistant to all antibiotics used in the study (58). In the study of Cherifi et al, 84 samples were isolated from hospitalized patients, 66 of which were S. epidermidis and the rest were S. aureus. Based on the results of the PCR test on S. epidermidis strains, 3 isolates had mecA. However, in the present study, out of 35 isolates of MRSA, only 4 isolates (11.4%) had the mecA gene, and out of 35 isolates of MRSE, 15 isolates (42.8%) had the mecA gene. The findings showed that the prevalence of the mecA gene among MRSA is decreasing, while the prevalence of this gene is increasing in MRSE (59). In a study performed by Noshak et al, S. epidermidis and S. haemolyticus strains were isolated from patients and healthcare workers. The detection of methicillin resistance among isolates was accomplished via the utilization of the cefoxitin disk diffusion test. Cefoxitin and cotrimoxazole demonstrated the highest resistance rates, with a value of 81.5%. Among all the MRSE and methicillin-resistant S. haemolyticus isolates, 66 mecA-positive isolates were detected (60). In further analysis, it was found that out of the 27 isolates of S. aureus, approximately 55.6% of the isolates exhibited MRSA. The PCR analysis involved choosing various strains of S. aureus for examination. It was observed that a substantial proportion (53.3%) of the MRSA isolates were found to possess the mecAgene. Conversely, it was noted that all MSSA isolates tested negative for the presence of the mecA gene (61). A study conducted by Siddiqui et al yielded similar findings, indicating that only 36.5% of the subjects exhibited mecApositivity. However, all MSSA isolates tested negative in PCR (62). Due to the high prevalence and clinical importance of these infections, it is necessary for the hospital staff to be aware and plan to develop methods of prevention, treatment, and successful control of these infections in treatment systems (infection control unit), especially for patients with immune system deficiency in the community and hospitals are a priority.
Conclusion
Evaluating the prevalence of isolates with virulence genes as well as investigating drug resistance in hospitals can be effective in controlling infectious diseases in people with immune system deficiencies. The present study found that the incidence of the mecA gene in MRSA is in decline, whereas it is on the rise for MRSE.
Acknowledgements
The authors of this article express gratitude towards the personnel of the microbiology department at Omid (Seyed Al-Shohdai) Hospital in Isfahan, as well as the staff and personnel of Taligene Pars Science Foundation located in Isfahan Scientific Research Town for their cooperation and assistance. Their provision of an appropriate space and place and valuable guidance was instrumental in the successful execution of microbiological and molecular tests.
Authors’ Contribution
Conceptualization: Zahra Babaei.
Data collection: Zahra Babaei.
Formal analysis: Zahra Babaei.
Funding acquisition: Zahra Babaei.
Investigation: Zahra Babaei.
Methodology: Zahra Babaei.
Project administration: Monir Doudi, Zahra Babaei, Ladan Rahimzadeh Torabi.
Resources: Zahra Babaei.
Software: Monir Doudi, Zahra Babaei, Ladan Rahimzadeh Torabi.
Supervision: Monir Doudi, Zahra Babaei.
Validation: Monir Doudi, Zahra Babaei, Ladan Rahimzadeh Torabi.
Competing Interests
There is no conflict of interests as stated by the authors.
Ethical Approval
Not applicable.
Funding
Not applicable.
References
- Severn MM, Horswill AR. Staphylococcus epidermidis and its dual lifestyle in skin health and infection. Nat Rev Microbiol 2023; 21(2):97-111. doi: 10.1038/s41579-022-00780-3 [Crossref] [ Google Scholar]
- Piruozi A, Forouzandeh H, Farahani A, Askarpour M, Mohseni P, Fariyabi F. Frequency of nosocomial bacterial infections in hospitalized patients referred to Amir Al-Momenin Hospital, Gerash, Iran. Gene Cell Tissue 2019; 6(3):e93160. doi: 10.5812/gct.93160 [Crossref] [ Google Scholar]
- Guo Y, Song G, Sun M, Wang J, Wang Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front Cell Infect Microbiol 2020; 10:107. doi: 10.3389/fcimb.2020.00107 [Crossref] [ Google Scholar]
- Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, Khoon LY, Aziz MN, Hamat RA. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol 2010; 48(3):867-72. doi: 10.1128/jcm.01112-09 [Crossref] [ Google Scholar]
- Forozeshfard M, Ghorbani R, Razavi M, Danaie N, Nooripour S. Comparison of the umbilical cord bacterial colonization in newborn infants rooming in with mothers and neonates admitted to neonatal intensive care unit. Int J Pediatr 2017; 5(11):6009-15. doi: 10.22038/ijp.2017.25938.2208 [Crossref] [ Google Scholar]
- Brazel M, Desai A, Are A, Motaparthi K. Staphylococcal scalded skin syndrome and bullous impetigo. Medicina (Kaunas) 2021; 57(11):1157. doi: 10.3390/medicina57111157 [Crossref] [ Google Scholar]
- Algammal AM, Hetta HF, Elkelish A, Alkhalifah DHH, Hozzein WN, Batiha GE. Methicillin-resistant Staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect Drug Resist 2020; 13:3255-65. doi: 10.2147/idr.s272733 [Crossref] [ Google Scholar]
- Mohammad Alam F, Tasnim T, Afroz S, Mohammad Alam AR, Afroze N, Khatun A. Epidemiology and antibiogram of clinical Staphylococcus aureus isolates from tertiary care hospitals in Dhaka, Bangladesh. Avicenna J Clin Microbiol Infect 2022; 9(4):137-47. doi: 10.34172/ajcmi.2022.3391 [Crossref] [ Google Scholar]
- Bæk KT, Gründling A, Mogensen RG, Thøgersen L, Petersen A, Paulander W. β-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob Agents Chemother 2014; 58(8):4593-603. doi: 10.1128/aac.02802-14 [Crossref] [ Google Scholar]
- Torabi LR, Naghavi NS, Doudi M, Monajemi R. Efficacious antibacterial potency of novel bacteriophages against ESBL-producing Klebsiella pneumoniae isolated from burn wound infections. Iran J Microbiol 2021; 13(5):678-690. doi: 10.18502/ijm.v13i5.7435 [Crossref] [ Google Scholar]
- Månsson E, Tevell S, Nilsdotter-Augustinsson Å, Johannesen TB, Sundqvist M, Stegger M. Methicillin-resistant Staphylococcus epidermidis lineages in the nasal and skin microbiota of patients planned for arthroplasty surgery. Microorganisms 2021; 9(2):265. doi: 10.3390/microorganisms9020265 [Crossref] [ Google Scholar]
- Chabi R, Momtaz H. Virulence factors and antibiotic resistance properties of the Staphylococcus epidermidis strains isolated from hospital infections in Ahvaz, Iran. Trop Med Health 2019; 47:56. doi: 10.1186/s41182-019-0180-7 [Crossref] [ Google Scholar]
- Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol 2019; 17(4):203-18. doi: 10.1038/s41579-018-0147-4 [Crossref] [ Google Scholar]
- Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 2006; 44(1):108-18. doi: 10.1128/jcm.44.1.108-118.2006 [Crossref] [ Google Scholar]
- Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 2018; 31(4):e00020-18. doi: 10.1128/cmr.00020-18 [Crossref] [ Google Scholar]
- Harkins CP, Pichon B, Doumith M, Parkhill J, Westh H, Tomasz A. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol 2017; 18(1):130. doi: 10.1186/s13059-017-1252-9 [Crossref] [ Google Scholar]
- Vestergaard M, Frees D, Ingmer H. Antibiotic resistance and the MRSA problem. Microbiol Spectr 2019;7(2). 10.1128/microbiolspec.GPP3-0057-2018.
- Shalaby MW, Dokla EME, Serya RAT, Abouzid KAM. Penicillin binding protein 2A: an overview and a medicinal chemistry perspective. Eur J Med Chem 2020; 199:112312. doi: 10.1016/j.ejmech.2020.112312 [Crossref] [ Google Scholar]
- Young M, Walsh DJ, Masters E, Gondil VS, Laskey E, Klaczko M. Identification of Staphylococcus aureus penicillin binding protein 4 (PBP4) inhibitors. Antibiotics (Basel) 2022; 11(10):1351. doi: 10.3390/antibiotics11101351 [Crossref] [ Google Scholar]
- Fishovitz J, Hermoso JA, Chang M, Mobashery S. Penicillin-binding protein 2A of methicillin-resistant Staphylococcus aureus. IUBMB Life 2014; 66(8):572-7. doi: 10.1002/iub.1289 [Crossref] [ Google Scholar]
- Ballhausen B, Kriegeskorte A, Schleimer N, Peters G, Becker K. The mecA homolog mecC confers resistance against β-lactams in Staphylococcus aureus irrespective of the genetic strain background. Antimicrob Agents Chemother 2014; 58(7):3791-8. doi: 10.1128/aac.02731-13 [Crossref] [ Google Scholar]
- Uehara Y. Current status of staphylococcal cassette chromosome mec (SCCmec). Antibiotics (Basel) 2022; 11(1):86. doi: 10.3390/antibiotics11010086 [Crossref] [ Google Scholar]
- Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, Staphylococcus cassette chromosome mec, encodes methicillin-resistance in Staphylococcus aureus. Antimicrob Agents Chemother 2000; 44(6):1549-55. doi: 10.1128/aac.44.6.1549-1555.2000 [Crossref] [ Google Scholar]
- Wielders CL, Fluit AC, Brisse S, Verhoef J, Schmitz FJ. mecA gene is widely disseminated in Staphylococcus aureus population. J Clin Microbiol 2002; 40(11):3970-5. doi: 10.1128/jcm.40.11.3970-3975.2002 [Crossref] [ Google Scholar]
- Garoy EY, Gebreab YB, Achila OO, Tekeste DG, Kesete R, Ghirmay R. Methicillin-resistant Staphylococcus aureus (MRSA): prevalence and antimicrobial sensitivity pattern among patients-a multicenter study in Asmara, Eritrea. Can J Infect Dis Med Microbiol 2019; 2019:8321834. doi: 10.1155/2019/8321834 [Crossref] [ Google Scholar]
- Lee AS, de Lencastre H, Garau J, Kluytmans J, Malhotra-Kumar S, Peschel A. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers 2018; 4:18033. doi: 10.1038/nrdp.2018.33 [Crossref] [ Google Scholar]
- Morikawa K, Okada F, Ando Y, Ishii R, Matsushita S, Ono A. Meticillin-resistant Staphylococcus aureus and meticillin-susceptible S aureus pneumonia: comparison of clinical and thin-section CT findings. Br J Radiol 2012; 85(1014):e168-75. doi: 10.1259/bjr/65538472 [Crossref] [ Google Scholar]
- Boucher H, Miller LG, Razonable RR. Serious infections caused by methicillin-resistant Staphylococcus aureus. Clin Infect Dis 2010; 51 Suppl 2:S183-97. doi: 10.1086/653519 [Crossref] [ Google Scholar]
- Mahajan SN, Shah JN, Hachem R, Tverdek F, Adachi JA, Mulanovich V. Characteristics and outcomes of methicillin-resistant Staphylococcus aureus bloodstream infections in patients with cancer treated with vancomycin: 9-year experience at a comprehensive cancer center. Oncologist 2012; 17(10):1329-36. doi: 10.1634/theoncologist.2012-0029 [Crossref] [ Google Scholar]
- Li Z, Zhuang H, Wang G, Wang H, Dong Y. Prevalence, predictors, and mortality of bloodstream infections due to methicillin-resistant Staphylococcus aureus in patients with malignancy: systemic review and meta-analysis. BMC Infect Dis 2021; 21(1):74. doi: 10.1186/s12879-021-05763-y [Crossref] [ Google Scholar]
- van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. Predictors of mortality in Staphylococcus aureus bacteremia. Clin Microbiol Rev 2012; 25(2):362-86. doi: 10.1128/cmr.05022-11 [Crossref] [ Google Scholar]
- DeLeo FR, Diep BA, Otto M. Host defense and pathogenesis in Staphylococcus aureus infections. Infect Dis Clin North Am 2009; 23(1):17-34. doi: 10.1016/j.idc.2008.10.003 [Crossref] [ Google Scholar]
- Hasanpour AH, Sepidarkish M, Mollalo A, Ardekani A, Almukhtar M, Mechaal A. The global prevalence of methicillin-resistant Staphylococcus aureus colonization in residents of elderly care centers: a systematic review and meta-analysis. Antimicrob Resist Infect Control 2023; 12(1):4. doi: 10.1186/s13756-023-01210-6 [Crossref] [ Google Scholar]
- Martínez-Santos VI, Torres-Añorve DA, Echániz-Aviles G, Parra-Rojas I, Ramírez-Peralta A, Castro-Alarcón N. Characterization of Staphylococcus epidermidis clinical isolates from hospitalized patients with bloodstream infection obtained in two time periods. PeerJ 2022; 10:e14030. doi: 10.7717/peerj.14030 [Crossref] [ Google Scholar]
- Cabrera-Contreras R, Santamaría RI, Bustos P, Martínez-Flores I, Meléndez-Herrada E, Morelos-Ramírez R. Genomic diversity of prevalent Staphylococcus epidermidis multidrug-resistant strains isolated from a children’s hospital in México City in an eight-years survey. PeerJ 2019; 7:e8068. doi: 10.7717/peerj.8068 [Crossref] [ Google Scholar]
- Siciliano V, Passerotto RA, Chiuchiarelli M, Leanza GM, Ojetti V. Difficult-to-treat pathogens: a review on the management of multidrug-resistant Staphylococcus epidermidis. Life (Basel) 2023; 13(5):1126. doi: 10.3390/life13051126 [Crossref] [ Google Scholar]
- Peixoto PB, Massinhani FH, Netto Dos Santos KR, Chamon RC, Silva RB, Lopes Correa FE. Methicillin-resistant Staphylococcus epidermidis isolates with reduced vancomycin susceptibility from bloodstream infections in a neonatal intensive care unit. J Med Microbiol 2020; 69(1):41-5. doi: 10.1099/jmm.0.001117 [Crossref] [ Google Scholar]
- Rahimzadeh Torabi L, Doudi M, Naghavi NS, Monajemi R. Isolation, characterization, and effectiveness of bacteriophage Pɸ-Bw-Ab against XDR Acinetobacter baumannii isolated from nosocomial burn wound infection. Iran J Basic Med Sci 2021; 24(9):1254-63. doi: 10.22038/ijbms.2021.57772.12850 [Crossref] [ Google Scholar]
- Hos NJ, Jazmati N, Stefanik D, Hellmich M, AlSael H, Kern WV. Determining vancomycin Etest MICs in patients with MRSA bloodstream infection does not support switching antimicrobials. J Infect 2017; 74(3):248-59. doi: 10.1016/j.jinf.2016.12.007 [Crossref] [ Google Scholar]
- Rahimzadeh Torabi L, Naghavi NS, Doudi M, Monajemi R. Efficacious antibacterial potency of novel bacteriophages against ESBL-producing Klebsiella pneumoniae isolated from burn wound infections. Iran J Microbiol 2021; 13(5):678-90. doi: 10.18502/ijm.v13i5.7435 [Crossref] [ Google Scholar]
- Liu Y, Zhang J, Ji Y. PCR-based approaches for the detection of clinical methicillin-resistant Staphylococcus aureus. Open Microbiol J 2016; 10:45-56. doi: 10.2174/1874285801610010045 [Crossref] [ Google Scholar]
- Otto M. Staphylococcus epidermidis--the ‘accidental’ pathogen. Nat Rev Microbiol 2009; 7(8):555-67. doi: 10.1038/nrmicro2182 [Crossref] [ Google Scholar]
- Chessa D, Ganau G, Spiga L, Bulla A, Mazzarello V, Campus GV. Staphylococcus aureus and Staphylococcus epidermidis virulence strains as causative agents of persistent infections in breast implants. PLoS One 2016; 11(1):e0146668. doi: 10.1371/journal.pone.0146668 [Crossref] [ Google Scholar]
- Navidinia M, Zamani S, Mohammadi A, Araghi S, Amini C, Pourhossein B. Hospital-related lineage of USA300 methicillin-resistant Staphylococcus aureus (MRSA) to cause bacteremia in Iran. Biomed Res Int 2023; 2023:8335385. doi: 10.1155/2023/8335385 [Crossref] [ Google Scholar]
- Morell EA, Balkin DM. Methicillin-resistant Staphylococcus aureus: a pervasive pathogen highlights the need for new antimicrobial development. Yale J Biol Med 2010; 83(4):223-33. [ Google Scholar]
- Pantosti A, Sanchini A, Monaco M. Mechanisms of antibiotic resistance in Staphylococcus aureus. Future Microbiol 2007; 2(3):323-34. doi: 10.2217/17460913.2.3.323 [Crossref] [ Google Scholar]
- Pishva E, Havaei SA, Arsalani F, Narimani T, Azimian A, Akbari M. Detection of methicillin-resistance gene in Staphylococcus epidermidis strains isolated from patients in Al-Zahra hospital using polymerase chain reaction and minimum inhibitory concentration methods. Adv Biomed Res 2013; 2:23. doi: 10.4103/2277-9175.108008 [Crossref] [ Google Scholar]
- Sharma V, Jindal N, Devi P. Prevalence of methicillin-resistant coagulase negative staphylococci in a tertiary care hospital. Iran J Microbiol 2010; 2(4):185-8. [ Google Scholar]
- Idrees MM, Saeed K, Shahid MA, Akhtar M, Qammar K, Hassan J. Prevalence of mecA- and mecC-associated methicillin-resistant Staphylococcus aureus in clinical specimens, Punjab, Pakistan. Biomedicines 2023; 11(3):878. doi: 10.3390/biomedicines11030878 [Crossref] [ Google Scholar]
- Jafari-Sales A, Jafari B. Evaluation of the prevalence of mecA gene in Staphylococcus aureus strains isolated from clinical specimens of hospitals and treatment centers. Pajouhan Sci J 2019; 17(3):41-7. doi: 10.52547/psj.17.3.41 [Crossref] [ Google Scholar]
- Zerehsaz J, Najar Pirayeh S. Prevalence of mecA, tsst1, and pvl, as well as agr specific groups in clinical isolates of Staphylococcus aureus from patients admitted to hospitals in Tehran, Iran. Qom Univ Med Sci J 2020; 14(9):59-68. doi: 10.52547/qums.14.9.59 [Crossref] [ Google Scholar]
- de Matos PD, Schuenck RP, Cavalcante FS, Caboclo RM, dos Santos KR. Accuracy of phenotypic methicillin susceptibility methods in the detection of Staphylococcus aureus isolates carrying different SCCmec types. Mem Inst Oswaldo Cruz 2010; 105(7):931-4. doi: 10.1590/s0074-02762010000700017 [Crossref] [ Google Scholar]
- Rahimi F, Bouzari M, Katouli M, Pourshafie M. Prophage typing of methicillin-resistant Staphylococcus aureus isolated from a tertiary care hospital in Tehran, Iran. Jundishapur J Microbiol 2012; 6(1):80-5. doi: 10.5812/jjm.4616 [Crossref] [ Google Scholar]
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 2007; 51(1):264-74. doi: 10.1128/aac.00165-06 [Crossref] [ Google Scholar]
- Leung F, Richards CJ, Garbuz DS, Masri BA, Duncan CP. Two-stage total hip arthroplasty: how often does it control methicillin-resistant infection?. Clin Orthop Relat Res 2011; 469(4):1009-15. doi: 10.1007/s11999-010-1725-6 [Crossref] [ Google Scholar]
- Prasad S, Nayak N, Satpathy G, Nag HL, Venkatesh P, Ramakrishnan S. Molecular & phenotypic characterization of Staphylococcus epidermidis in implant related infections. Indian J Med Res 2012; 136(3):483-90. [ Google Scholar]
- Ternes YM, Lamaro-Cardoso J, André MC, Pessoa VP Jr, Vieira MA, Minamisava R. Molecular epidemiology of coagulase-negative Staphylococcus carriage in neonates admitted to an intensive care unit in Brazil. BMC Infect Dis 2013; 13:572. doi: 10.1186/1471-2334-13-572 [Crossref] [ Google Scholar]
- Du X, Zhu Y, Song Y, Li T, Luo T, Sun G. Molecular analysis of Staphylococcus epidermidis strains isolated from community and hospital environments in China. PLoS One 2013; 8(5):e62742. doi: 10.1371/journal.pone.0062742 [Crossref] [ Google Scholar]
- Cherifi S, Byl B, Deplano A, Nagant C, Nonhoff C, Denis O. Genetic characteristics and antimicrobial resistance of Staphylococcus epidermidis isolates from patients with catheter-related bloodstream infections and from colonized healthcare workers in a Belgian hospital. Ann Clin Microbiol Antimicrob 2014; 13:20. doi: 10.1186/1476-0711-13-20 [Crossref] [ Google Scholar]
- Noshak MA, Ahangarzadeh Rezaee M, Hasani A, Mirzaii M, Memar MY, Azimi T. Molecular detection and characterization of the Staphylococcus epidermidis and Staphylococcus haemolyticus isolated from hospitalized patients and healthcare workers in Iran. Biomed Res Int 2023; 2023:3775142. doi: 10.1155/2023/3775142 [Crossref] [ Google Scholar]
- Gaire U, Thapa Shrestha U, Adhikari S, Adhikari N, Bastola A, Rijal KR. Antibiotic susceptibility, biofilm production, and detection of mecA gene among Staphylococcus aureus isolates from different clinical specimens. Diseases 2021; 9(4):80. doi: 10.3390/diseases9040080 [Crossref] [ Google Scholar]
- Siddiqui T, Muhammad IN, Khan MN, Fatima S, Alam N, Masood R. Prevalence of mecA: genotyping screening of community acquired-MRSA isolates in Karachi, Pakistan. Pak J Pharm Sci 2018; 31(5):2091-4. [ Google Scholar]