Avicenna Journal of Clinical Microbiology and Infection. 10(3):100-105.
doi: 10.34172/ajcmi.3450
Original Article
Effect of Selenium Nanoparticles on the Expression of OqxB Gene in Clinical Isolates of Klebsiella pneumoniae
Wassen Hamid Abdolmasoudi 1  , Ashraf Kariminik 2, *
, Ashraf Kariminik 2, *  , Atousa Ferdousi 3
, Atousa Ferdousi 3 
Author information:
1Department of Microbiology, Science and Research Branch, Islamic Azad University, Tehran, Iran
2Department of Microbiology, Kerman Branch, Islamic Azad University, Kerman, Iran
3Department of Microbiology, Shahr-e-Qods Branch, Islamic Azad University, Tehran, Iran
Abstract
Background: OqxB is an efflux pump that has emerged as a factor contributing to antibiotic resistance in Klebsiella pneumoniae. The objective of this study was to investigate the occurrence of AcrAB efflux pump resistance genes in clinical samples of K. pneumoniae and to evaluate the influence of selenium nanoparticles (Se-NPs), loaded with ampicillin, on the expression of the OqxB gene associated with efflux pumps.
Methods: A total of 500 clinical samples were collected from hospitalized patients, and 60 strains of K. pneumoniae were isolated using standard microbiological methods. These strains were then analyzed using phenotypic and polymerase chain reaction (PCR) techniques to detect the frequency of KPSM, MRK, OqxA, and OqxB genes through multiplex PCR. The impact of Se-NPs loaded with ampicillin on the expression of the OqxB resistance gene was investigated using a real-time PCR technique.
Results: Based on the results of this study, it was found that the KPSM gene is not present in any of the investigated K. pneumoniae isolates. However, the MRK, OqxA, and OqxB genes were detected in 57, 55, and 54 isolates, respectively. The minimum inhibitory concentration (MIC) values for Se-NP and Se-NPs with ampicillin were reported to be 1500 μg/mL and 375 μg/mL, respectively. Notably, the SeNPs with ampicillin could significantly down-regulate the expression of the OqxB gene. These findings demonstrated the potential of Se-NPs as a promising strategy for reducing antibiotic resistance in K. pneumoniae infections.
Conclusion: The findings emphasize the notable prevalence of drug resistance genes, specifically those associated with efflux pump production, in clinical samples of K. pneumoniae. Remarkably, the utilization of Se nanoparticles loaded with ampicillin demonstrated its efficacy in suppressing the expression of the OqxB gene and enhancing bacterial susceptibility to ampicillin. The results further imply that Se-NPs could serve as a promising avenue for the development of innovative antibacterial agents, aimed at combating antibiotic resistance in K. pneumoniae infections.
Keywords: Klebsiella pneumoniae, Efflux pumps, PCR, Selenium nanoparticles
Copyright and License Information
© 2023 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
https://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Please cite this article as follows: Abdolmasoudi WH, Kariminik A, Ferdousi A. Effect of selenium nanoparticles on the expression of OqxB gene in clinical isolates of Klebsiella pneumoniae. Avicenna J Clin Microbiol Infect. 2023; 10(3):100-105. doi:10.34172/ajcmi.3450
Introduction
The development of multidrug-resistant bacterial pathogens is a major global public health challenge (1). Antibacterial resistance rates are high in several bacterial pathogens, especially Enterobacteriaceae, which limits therapeutic options for treating serious infections (2). Klebsiella pneumoniae is one of the multidrug-resistant organisms that poses a significant threat to human health in both community and hospital settings (3). This bacterium causes a wide range of infections, including septicemia, pneumonia, urinary tract infection, meningitis, and purulent abscesses in different organs, particularly in liver abscesses (4). The production of biofilm, especially in medical devices, and its association with increased horizontal transfer of antibiotic resistance genes is a significant factor that complicates efforts to control these infections. Isolates’ ability to create biofilm results in an increase in antibiotic resistance by more than a hundred times (5).
Multidrug-resistant pumps are commonly found in bacterial chromosomes, providing them with an intrinsic ability to develop resistance without the need for acquiring antibiotic resistance genes. K. pneumoniae is known to express the AcrAB efflux pump, which facilitates not only the efflux of antibiotics such as quinolones and β-lactams but also host-derived antimicrobial agents such as those found in human alveolar bronchoalveolar lavage fluid and antimicrobial peptides (6). Consequently, AcrAB serves as a resistance determinant against the host’s innate immune defense system in K. pneumoniae (7).
The resistance mechanism includes the AcrAB multidrug efflux system, which is responsible for this phenotype in K. pneumoniae. The acrRAB operon encodes various components of this system. Within the operon, acrR codes for the AcrAB repressor, while acrA and acrB encode a periplasmic lipoprotein weighing 40 kDa, which is anchored to the inner membrane and acts as a bridge between the outer and inner membranes and an integral membrane protein weighing 113.5 kDa. The integral membrane protein consists of 12 membrane-spanning α-helices and is located in the cytoplasmic membrane (8).
OqxB belongs to the resistance-nodulation-division efflux pump family and has been identified as a key contributor to resistance against different antibiotics, including quinolones, nitrofurantoin, quinoxalines, tigecycline, and chloramphenicol, as well as detergents and disinfectants. While there are reports on the presence of oqxB and its associated resistance, limited information is available regarding its other characteristics or mechanisms of action (9). Selenium (Se) is a trace element essential to the human body due to its antioxidant and pro-oxidative effects. The recommended daily intake of Se is approximately 40 μg, but high doses of up to 400 μg / day can be toxic (10). Se plays a vital role in biological functions and acts as an important cofactor for antioxidant enzymes such as glutathione peroxidases and thioredoxin reductases that protect against free radical species (11). Recently, there has been a growing interest in the preparation and study of Se nanoparticles (Se-NPs) due to their interesting biological activities in both in vitro and in vivo conditions. Se-NPs exhibit low toxicity and excellent bioavailability compared to Se, making them an attractive alternative for various biomedical applications (12).
This study aimed to investigate the prevalence of AcrAB efflux pump genes in clinical isolates of K. pneumoniae as well as the impact of Se-NPs on the survival of K. pneumoniae and the expression of an important member of this drug resistance gene, the OqxB gene.
Materials and Methods
Isolation and Identification of Klebsiella pneumoniae Isolates
In this descriptive cross-sectional study, a total of 500 clinical samples (e.g., blood, urine, and wound) were collected from patients admitted to hospitals in Tehran between February and September 2022. The K. pneumoniae isolates were identified using conventional diagnostic tests (13,14). The isolated bacteria were stored at -20°C in brain heart infusion broth containing 20% glycerol for further investigation. To confirm the isolates, a specific target was amplified using the tyrB gene and conventional polymerase chain reaction (PCR). Genomic DNA was extracted using a genomic DNA extraction kit (CinnaGen Company, Iran) according to the manufacturer’s instructions. The PCR mixture contained 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 1 μL purified DNA (1 μg), 10 μL PCR master mix (Fermentase Company, Germany), and 7 μL DNase/RNase free water. The primer sequence of the tyrB gene is listed in Table 1. The cycling conditions included an initial denaturation for 5 minutes at 95 °C, 40 cycles of 1 minute at 95 °C, 1 minute at 55 °C, and 1 minute at 72 °C, and a final extension for 10 minutes at 72 °C. The PCR product was then separated on a 2% agarose gel and stained with ethidium bromide (15).
Table 1.
Genes and Primer Sequences
|
Gene
|
Sequences
|
Size (bp)
|
Reference
|
|
tyrB
|
F5'-GGCTGTACTACAACGATGAC -3`
R5'-TTGAGCAGGTAATCCACTTTG -3' |
931 |
(15) |
|
MRK
|
F5'-GTCTTTTCGTCCCGGGTATATAAC-3'
R5'-CCACATCGACATTCATATTTTTCC-3' |
244 |
(16) |
|
KPSM
|
F5'-GCGCATTTGCTGATACTGTTG-3'
R5'-CATCAGACGATAAGCATGAGC-3' |
272 |
(17) |
|
OqxA
|
F5'-CGTGCTGTTCACGATAGATG-3'
R5'-GACACGAGGTTGGTATGGAC-3' |
144 |
This study |
|
OqxB
|
F5'-CGGCCAGTTCTACAAACAGT-3'
R5'-GGTAGGGAGGTCTTTCTTCG-3' |
136 |
(18) |
Polymerase Chain Reaction Detection of Efflux Pump Genes
Multiplex PCR was employed to detect the frequency of KPSM, MRK, OqxA, and OqxB genes. The PCR conditions included an initial denaturing at 94 °C for 5 minutes, followed by 35 cycles consisting of 1 minute at 94 °C for denaturation, 1 minute at 58 °C, 60 seconds for extension steps, and finally one cycle for the final extension at 72°C for 10 minutes. The primers used for the target genes are listed in Table 1.
Synthesis of Selenium Nanoparticles
The Se-NPs were synthesized through the chemical reduction of sodium selenite salt with L-cysteine amino acid. To achieve this, a solution of sodium selenite salt was prepared along with a L-cysteine solution. Subsequently, 10 mL of the 0.1 molar sodium selenite was transferred to a volumetric flask (1000 mL), and 40 mL of a 50 mM L-cysteine solution was added drop by drop to the flask using a graduated pipette. The volume was then increased to 1000 mL by adding distilled water. The mixture was continuously stirred at 6000 rpm for 30 minutes at 25 °C, resulting in the formation of a red-to-brick colloidal solution of Se-NPs. The NPs were dried at a temperature of 25-30 °C, and the resulting dried NP powder was collected and stored for further studies. The NPs were characterized using dynamic light scattering (DLS), zeta potential, and scanning electron microscopy (SEM).
Ampicillin Loading on Selenium Nanoparticles
To prepare the Se-NPs for further studies, 50 mg of the NPs were added to a 25 mL flask containing 10 mL of deionized distilled water. The mixture was then placed in an ultrasonic bath with medium speed for 10 minutes to ensure complete dispersion of the NPs in the water. Ampicillin antibiotic solution (Sigma) with a concentration of 50 μg/mL was added drop by drop into the container containing the dispersed nanoparticles, reaching a final volume of 25 mL. The mixture was then placed in a shaker incubator at a speed of 150 rpm and a temperature of 25 °C for 24 hours. After this period, the contents of the flask were centrifuged at 13 000 rpm for 15-20 minutes, and the resulting precipitate was collected for further analysis.
Effects of Ampicillin/Selenium Nanoparticles on the Expression of OqxB
The real-time PCR technique was employed to determine the effects of Se-NPs on the expression of the OqxBgene, which is an important resistant gene of efflux pump genes. To evaluate the impact of ampicillin/Se-NPs on the expression levels of OqxB, the bacteria were cultured in sub-minimum inhibitory concentrations (MICs) of the ampicillin/Se-NPs, and after 24 hours of incubation at 37 °C, total RNA was extracted using a commercial kit (Fermentas Company, Germany). The concentration and purity of the extracted RNA were evaluated at 260 and 280 nm. The purified RNA was then converted into cDNA using a commercial kit from Fermentas Company, Germany, following the manufacturer’s instructions. Then, real-time PCR was performed using a Step-One PLUS ABI thermocycler (USA). A mixture containing 2 μL of primers (10 mM, Table 1), 10 μL SYBR Green master mix (CinnaGen Company, Iran), 1.5 μL cDNA (1 mg), and 6.5 μL RNase/DNase free water was added to special microtubes. The expression of K. pneumoniae 16s rRNA was measured as the housekeeping gene, and the 2-∆∆Ct formula was used to normalize the raw data.
Statistical Analysis
SPSS software version 21 was used for statistical analysis of the data. The Kolmogorov-Smirnov test was employed to explore the normality of the data distribution. One-way ANOVA was then used for statistical analysis of the variables, and the data were presented as mean ± standard error. Further, a P value less than 0.05 was considered statistically significant.
Results
In this study, a total of 60 isolates of K. pneumoniae were identified from clinical samples using biochemical and molecular methods. The results of the multiplex-PCR test for the efflux pump genes among the isolates revealed that none of the isolates have the KPSM gene. However, the MRK, OqxA, andOqxB genes were detected in 57, 55, and 54 isolates, respectively.
Characterization of Nanoparticles
According to the SEM evaluation, the Se-NPs exhibited a spherical morphology and had an average size of 53 nm (Figure 1). The hydrodynamic diameter and Zeta potential of the particles were also determined using a Zetasizer Nano. The determination of the Zeta potential was used to predict the stability of dispersed particles in an aqueous medium. The higher the absolute value of Zeta potential, the lower the likelihood of Se-NPs to aggregate. The average zeta potential values of the NPs ranged from 0 to 100 mV. On the other hand, DLS measurements of the NPs generated values with a hydrodynamic diameter of 8 nm (Figure 2).
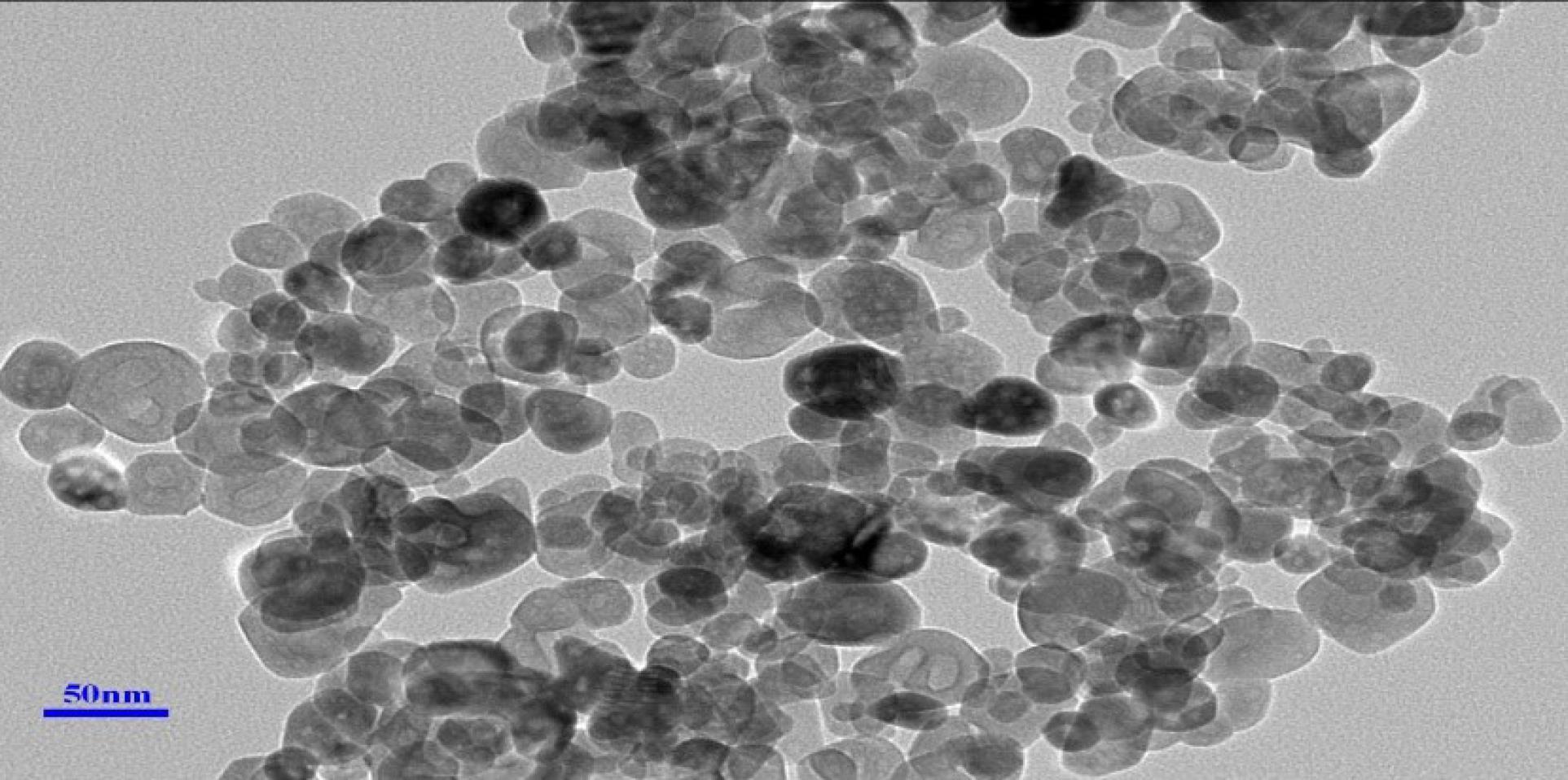
Figure 1.
SEM Image of Se Nanoparticles. Note. SEM: Scanning electron microscopy; Se: Selenium
.
SEM Image of Se Nanoparticles. Note. SEM: Scanning electron microscopy; Se: Selenium
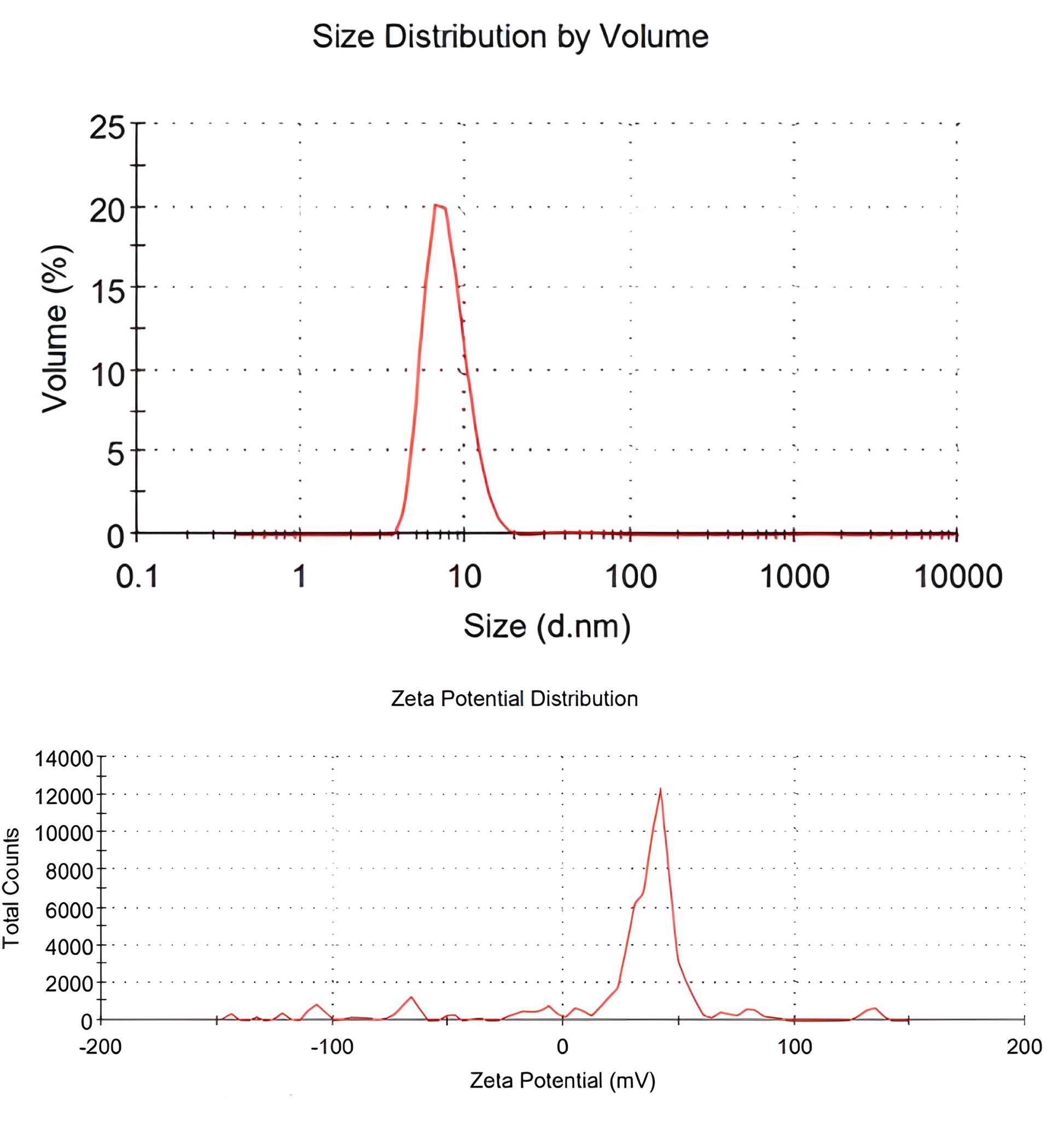
Figure 2.
Zeta Potential and Hydrodynamic Diameter of the Se Nanoparticles. Note. Se: Selenium
.
Zeta Potential and Hydrodynamic Diameter of the Se Nanoparticles. Note. Se: Selenium
Determination of the Minimum Inhibitory Concentration
The MIC and sub-MIC values for Se-NPs in this study were 1500 and 750 μg/mL, respectively. Meanwhile, the MIC and sub-MIC values for ampicillin/Se-NPs were determined to be 375 and 187.5 μg/mL, respectively.
The Effects of Selenium Nanoparticles on the Expression of OqxB Gene
The real-time PCR results revealed that the expression levels of the OqxB gene significantly decreased after treatment with ampicillin/Se-NPs (P = 0.020). Specifically, the expression levels of the gene were 1 ± 0.10 before treatment and 0.43 ± 0.05 after treatment with ampicillin/Se-NPs (Figure 3).
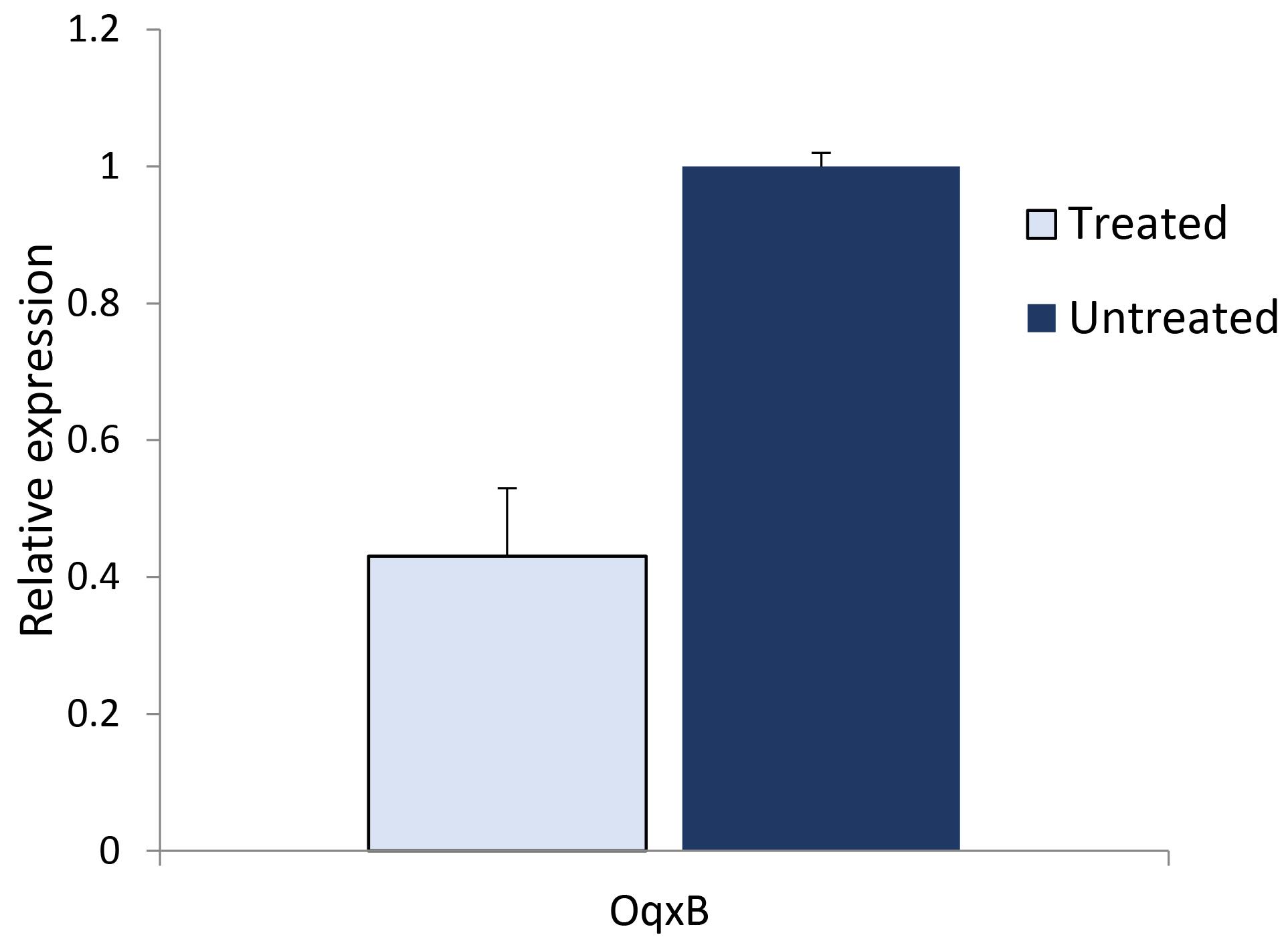
Figure 3.
Expression Levels of the OqxB Gene before and after Treatment by the Se Nanoparticles. Note. Se: Selenium
.
Expression Levels of the OqxB Gene before and after Treatment by the Se Nanoparticles. Note. Se: Selenium
Discussion
The global challenge of antibiotic resistance, particularly in the context of K. pneumoniae infections, has become a growing concern. This has increased mortality rates associated with infections caused by resistant strains of K. pneumoniae (19,20). As such, the World Health Organization (WHO) has designated K. pneumoniae as a priority pathogen, highlighting the urgent need for the development of next-generation antibiotics (21). Antibiotics that were previously effective against K. pneumoniae such as nitrofurantoin, beta-lactams, aminoglycosides, quinolones, tigecycline, and colistin, have now been shown to be ineffective due to the increased expression of efflux pumps (22). Most Gram-negative bacteria have multiple families of efflux pumps that reduce the intracellular concentration of antibiotics, resulting in acquired intrinsic resistance to several classes of antibiotics (23). However, the prevalence of these resistance genes varies among different ethnic populations. The current study demonstrated a high prevalence of resistance genes among patients infected with K. pneumoniae, indicating that treatment of these infections can be challenging and pose a significant risk to this population. These findings are consistent with those of Muhsin et al, who reported a high prevalence of efflux pump resistance genes among clinical isolates of K. pneumoniae in Baghdad, Iraq (24). Similar results were also observed in other studies conducted in different parts of the world (25,26). Similar to our research findings, Rodríguez-Martínez et al also discovered a high prevalence of oqxA and oqxB genes in K. pneumoniae, with rates of 76% and 75%, respectively (27). In a separate study, oqxA and oqxB genes were detected in 56.7% and 54.6% of K. pneumoniae isolates, respectively (28(. According to the study conducted by Yuan et al, both oqxA and oqxB genes were found to be present in all strains of K. pneumoniae that were analyzed (29). The increased prevalence of efflux pump resistance genes among K. pneumoniae isolates from patients is concerning and calls for urgent attention to identify new antibacterial strategies. Our findings suggest that ampicillin/Se-NPs may serve as a potential antibacterial agent as they could significantly reduce the resistance of K. pneumoniae to ampicillin and decrease the expression of the OqxB gene, which is a major antibiotic resistance gene.
This study revealed a noteworthy decrease in the expression of the OqxB gene, which could potentially play a role in diminishing resistance to various antibiotics. Moreover, the findings indicated that the utilization of ampicillin/Se-NPs can enhance the effectiveness of ampicillin and make bacteria more susceptible to the antibiotic. This enhanced sensitivity may be attributed to mechanisms that do not rely on the decreased expression of the OqxB gene. Several potential mechanisms could explain the increased sensitivity to ampicillin when utilizing ampicillin/Se-NPs (30). The NPs may improve the targeted delivery of ampicillin to the bacterial cells, ensuring that a higher concentration of the antibiotic reaches its intended target. This increased delivery can enhance the bactericidal effect of ampicillin. On the other hand, the NPs may facilitate the uptake of ampicillin by bacterial cells. They can interact with the cell membrane, promoting the entry of ampicillin into the cells and increasing its intracellular concentration. This higher concentration can lead to improved antibiotic efficacy. Furthermore, the combination of ampicillin with Se-NPs may exhibit synergistic effects, where the NPs enhance the antimicrobial activity of ampicillin. This synergy could involve various mechanisms such as disrupting bacterial cell walls or interfering with bacterial metabolic pathways. Additionally, the inclusion of ampicillin/Se-NPs in the system can potentially elicit cellular stress and inflict damage upon bacterial cells. These NPs can generate reactive oxygen species or disrupt vital cellular processes. As a result, bacteria may become more susceptible to the effects of ampicillin, thereby increasing their vulnerability to the antibiotic’s action (30,31). It is worth noting that these are hypothetical mechanisms, and further research is needed to determine the exact mechanisms responsible for the increased sensitivity to ampicillin when using ampicillin/Se-NPs. These findings are supported by a study by Huang et al who reported that Se-NPs coated with the antimicrobial polypeptide ε-poly-L-lysine (ε-PL) exhibit significant antibacterial activity against various strains of bacteria, including Gram-positive, Gram-negative, and drug-resistant strains compared to the individual components of Se-NPs and ε-PL (32). The promising therapeutic potential of Se-NPs was demonstrated in the current study as the NPs exhibited no toxicity to human dermal fibroblasts at minimal inhibitory concentrations. These findings suggest that the combination of Se-NPs with antibiotics and antimicrobial peptides may represent a highly effective new strategy with broad antibacterial activity, low cytotoxicity, and a significant delay in the development of resistance (32). The ability of Se-NPs to decrease antibiotic resistance in various bacteria has been demonstrated in previous studies (33-36), highlighting their potential as a valuable tool in the fight against antibiotic-resistant infections.
Conclusion
Our findings indicate that the utilization of Se-NPs in antibiotic preparation holds great promise in addressing drug resistance in different bacteria, particularly those associated with hospital-acquired infections. However, further research is needed to assess the impact of Se-NPs conjugated with other antibiotics on resistant bacteria as well as to investigate the potential influence of this novel drug on the expression of other resistance genes. When using Se-NPs in antibiotic preparation, several potential challenges or limitations may arise. The safety profile of Se-NPs needs to be thoroughly evaluated as their potential toxicity and long-term effects on human health and the environment have not yet been fully understood. It is crucial to ensure that the use of Se-NPs is safe for both patients and the environment. Additionally, NPs such as Se-NPs may exhibit instability over time, leading to changes in their physicochemical properties and potentially affecting their efficacy. Hence, ensuring the stability and shelf-life of Se-NPs in antibiotic formulations is essential for their practical application. Furthermore, the use of NPs in medical applications may require regulatory approvals and compliance with specific guidelines. Accordingly, adequate regulatory measures need to be in place to ensure the safe and ethical use of Se-NPs in antibiotic formulations.
Acknowledgements
The authors express their sincere gratitude to the research laboratory of the Science and Research Branch, Islamic Azad University of Tehran, Iran, for their guidance and valuable advice throughout the study. Their assistance has been instrumental in facilitating the successful completion of this research project.
Authors’ Contribution
Conceptualization: Wassen Hamid Abdolmasoudi.
Data Collection: Wassen Hamid Abdolmasoudi.
Formal analysis: Wassen Hamid Abdolmasoudi, Atousa Ferdousi.
Investigation: Wassen Hamid Abdolmasoudi.
Methodology: Wassen Hamid Abdolmasoudi, Ashraf Kariminik.
Validation: Ashraf Kariminik, Atousa Ferdousi.
Writing–original draft: Ashraf Kariminik.
Competing Interests
There is no conflict of interests as stated by the authors.
Ethical Approval
Informed consent was not required for this study.
References
- Kidd TJ, Mills G, Sá-Pessoa J, Dumigan A, Frank CG, Insua JL. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 2017; 9(4):430-47. doi: 10.15252/emmm.201607336 [Crossref] [ Google Scholar]
- Wang J, Wu H, Mei CY, Wang Y, Wang ZY, Lu MJ. Multiple mechanisms of tigecycline resistance in Enterobacteriaceae from a pig farm, China. Microbiol Spectr 2021; 9(2):e0041621. doi: 10.1128/Spectrum.00416-21 [Crossref] [ Google Scholar]
- Hawkey J, Cottingham H, Tokolyi A, Wick RR, Judd LM, Cerdeira L. Linear plasmids in Klebsiella and other Enterobacteriaceae. Microb Genom 2022; 8(4):000807. doi: 10.1099/mgen.0.000807 [Crossref] [ Google Scholar]
- Dong N, Yang X, Chan EW, Zhang R, Chen S. Klebsiella species: taxonomy, hypervirulence and multidrug resistance. EBioMedicine 2022; 79:103998. doi: 10.1016/j.ebiom.2022.103998 [Crossref] [ Google Scholar]
- Wang G, Zhao G, Chao X, Xie L, Wang H. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. Int J Environ Res Public Health 2020; 17(17):6278. doi: 10.3390/ijerph17176278 [Crossref] [ Google Scholar]
- Mirzaie A, Ranjbar R. Antibiotic resistance, virulence-associated genes analysis and molecular typing of Klebsiella pneumoniae strains recovered from clinical samples. AMB Express 2021; 11(1):122. doi: 10.1186/s13568-021-01282-w [Crossref] [ Google Scholar]
- Padilla E, Llobet E, Doménech-Sánchez A, Martínez-Martínez L, Bengoechea JA, Albertí S. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 2010; 54(1):177-83. doi: 10.1128/aac.00715-09 [Crossref] [ Google Scholar]
- Ni RT, Onishi M, Mizusawa M, Kitagawa R, Kishino T, Matsubara F. The role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae. Sci Rep 2020; 10(1):10876. doi: 10.1038/s41598-020-67820-x [Crossref] [ Google Scholar]
- Bharatham N, Bhowmik P, Aoki M, Okada U, Sharma S, Yamashita E. Structure and function relationship of OqxB efflux pump from Klebsiella pneumoniae. Nat Commun 2021; 12(1):5400. doi: 10.1038/s41467-021-25679-0 [Crossref] [ Google Scholar]
- Zhang JS, Gao XY, Zhang LD, Bao YP. Biological effects of a nano red elemental selenium. Biofactors 2001; 15(1):27-38. doi: 10.1002/biof.5520150103 [Crossref] [ Google Scholar]
- Carlisle AE, Lee N, Matthew-Onabanjo AN, Spears ME, Park SJ, Youkana D. Selenium detoxification is required for cancer-cell survival. Nat Metab 2020; 2(7):603-11. doi: 10.1038/s42255-020-0224-7 [Crossref] [ Google Scholar]
- Khurana A, Tekula S, Saifi MA, Venkatesh P, Godugu C. Therapeutic applications of selenium nanoparticles. Biomed Pharmacother 2019; 111:802-12. doi: 10.1016/j.biopha.2018.12.146 [Crossref] [ Google Scholar]
- Berkowitz FE, Jerris RC. Practical Medical Microbiology for Clinicians. John Wiley & Sons; 2016.
- Hansen DS, Aucken HM, Abiola T, Podschun R. Recommended test panel for differentiation of Klebsiella species on the basis of a trilateral interlaboratory evaluation of 18 biochemical tests. J Clin Microbiol 2004; 42(8):3665-9. doi: 10.1128/jcm.42.8.3665-3669.2004 [Crossref] [ Google Scholar]
- Jeong ES, Lee KS, Heo SH, Seo JH, Choi YK. Rapid identification of Klebsiella pneumoniae, Corynebacterium kutscheri, and Streptococcus pneumoniae using triplex polymerase chain reaction in rodents. Exp Anim 2013; 62(1):35-40. doi: 10.1538/expanim.62.35 [Crossref] [ Google Scholar]
- Bakhtiari R, Javadi A, Aminzadeh M, Molaee-Aghaee E, Shaffaghat Z. Association between presence of RmpA, MrkA and MrkD genes and antibiotic resistance in clinical Klebsiella pneumoniae isolates from hospitals in Tehran, Iran. Iran J Public Health 2021; 50(5):1009-16. doi: 10.18502/ijph.v50i5.6118 [Crossref] [ Google Scholar]
- Mamlouk K, Boutiba-Ben Boubaker I, Gautier V, Vimont S, Picard B, Ben Redjeb S. Emergence and outbreaks of CTX-M beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae strains in a Tunisian hospital. J Clin Microbiol 2006; 44(11):4049-56. doi: 10.1128/jcm.01076-06 [Crossref] [ Google Scholar]
- Razavi S, Mirnejad R, Babapour E. Involvement of AcrAB and OqxAB efflux pumps in antimicrobial resistance of clinical isolates of Klebsiella pneumonia. J Appl Biotechnol Rep 2020; 7(4):251-7. doi: 10.30491/jabr.2020.120179 [Crossref] [ Google Scholar]
- Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health 2015; 109(7):309-18. doi: 10.1179/2047773215y.0000000030 [Crossref] [ Google Scholar]
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 2017; 41(3):252-75. doi: 10.1093/femsre/fux013 [Crossref] [ Google Scholar]
- Kaczor AA, Polski A, Sobótka-Polska K, Pachuta-Stec A, Makarska-Bialokoz M, Pitucha M. Novel antibacterial compounds and their drug targets - successes and challenges. Curr Med Chem 2017; 24(18):1948-82. doi: 10.2174/0929867323666161213102127 [Crossref] [ Google Scholar]
- Lv L, Wan M, Wang C, Gao X, Yang Q, Partridge SR. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio 2020; 11(2):e02930-19. doi: 10.1128/mBio.02930-19 [Crossref] [ Google Scholar]
- Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev 2015; 28(2):337-418. doi: 10.1128/cmr.00117-14 [Crossref] [ Google Scholar]
- Muhsin EA, Sajid Al-Jubori S, Abdulhemid Said L. Prevalence of efflux pump and porin-related antimicrobial resistance in clinical Klebsiella pneumoniae in Baghdad, Iraq. Arch Razi Inst 2022; 77(2):785-98. doi: 10.22092/ari.2022.356976.1952 [Crossref] [ Google Scholar]
- Kareem SM, Al-Kadmy IMS, Kazaal SS, Mohammed Ali AN, Aziz SN, Makharita RR. Detection of gyrA and parC mutations and prevalence of plasmid-mediated quinolone resistance genes in Klebsiella pneumoniae. Infect Drug Resist 2021; 14:555-63. doi: 10.2147/idr.s275852 [Crossref] [ Google Scholar]
- Ahmed Hasan S, Fakhraddin Raheem T, Mohammed Abdulla H. Phenotypic, antibiotyping, and molecular detection of Klebsiella Pneumoniae isolates from clinical specimens in Kirkuk, Iraq. Arch Razi Inst 2021; 76(4):1061-7. doi: 10.22092/ari.2021.355770.1721 [Crossref] [ Google Scholar]
- Rodríguez-Martínez JM, Díaz de Alba P, Briales A, Machuca J, Lossa M, Fernández-Cuenca F. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemother 2013; 68(1):68-73. doi: 10.1093/jac/dks377 [Crossref] [ Google Scholar]
- Goudarzi M, Azad M, Seyedjavadi SS. Prevalence of plasmid-mediated quinolone resistance determinants and OqxAB efflux pumps among extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from patients with nosocomial urinary tract infection in Tehran, Iran. Scientifica (Cairo) 2015; 2015:518167. doi: 10.1155/2015/518167 [Crossref] [ Google Scholar]
- Yuan J, Xu X, Guo Q, Zhao X, Ye X, Guo Y. Prevalence of the oqxAB gene complex in Klebsiella pneumoniae and Escherichia coli clinical isolates. J Antimicrob Chemother 2012; 67(7):1655-9. doi: 10.1093/jac/dks086 [Crossref] [ Google Scholar]
- Khorsandi K, Hosseinzadeh R, Sadat Esfahani H, Keyvani-Ghamsari S, Ur Rahman S. Nanomaterials as drug delivery systems with antibacterial properties: current trends and future priorities. Expert Rev Anti Infect Ther 2021; 19(10):1299-323. doi: 10.1080/14787210.2021.1908125 [Crossref] [ Google Scholar]
- Canaparo R, Foglietta F, Limongi T, Serpe L. Biomedical applications of reactive oxygen species generation by metal nanoparticles. Materials (Basel) 2020; 14(1):53. doi: 10.3390/ma14010053 [Crossref] [ Google Scholar]
- Huang T, Holden JA, Reynolds EC, Heath DE, O’Brien-Simpson NM, O’Connor AJ. Multifunctional antimicrobial polypeptide-selenium nanoparticles combat drug-resistant bacteria. ACS Appl Mater Interfaces 2020; 12(50):55696-709. doi: 10.1021/acsami.0c17550 [Crossref] [ Google Scholar]
- Han HW, Patel KD, Kwak JH, Jun SK, Jang TS, Lee SH. Selenium nanoparticles as candidates for antibacterial substitutes and supplements against multidrug-resistant bacteria. Biomolecules 2021; 11(7):1028. doi: 10.3390/biom11071028 [Crossref] [ Google Scholar]
- Lin A, Liu Y, Zhu X, Chen X, Liu J, Zhou Y. Bacteria-responsive biomimetic selenium nanosystem for multidrug-resistant bacterial infection detection and inhibition. ACS Nano 2019; 13(12):13965-84. doi: 10.1021/acsnano.9b05766 [Crossref] [ Google Scholar]
- AlMatar M, Makky EA, Var I, Koksal F. The role of nanoparticles in the inhibition of multidrug-resistant bacteria and biofilms. Curr Drug Deliv 2018; 15(4):470-84. doi: 10.2174/1567201815666171207163504 [Crossref] [ Google Scholar]
- Huang YS, Wang JT, Tai HM, Chang PC, Huang HC, Yang PC. Metal nanoparticles and nanoparticle composites are effective against Haemophilus influenzae, Streptococcus pneumoniae, and multidrug-resistant bacteria. J Microbiol Immunol Infect 2022; 55(4):708-15. doi: 10.1016/j.jmii.2022.05.003 [Crossref] [ Google Scholar]