Avicenna Journal of Clinical Microbiology and Infection. 9(4):152-156.
doi: 10.34172/ajcmi.2022.3402
Original Article
Isolation and Identification of Salmonella Typhimurium in Retail Meat Sources by Multiplex Polymerase Chain Reaction
Mazen Safi *  , Bassam Al Balaa , Samah Qasem , Laila Al Hallab , Ayman Al-Mariri
, Bassam Al Balaa , Samah Qasem , Laila Al Hallab , Ayman Al-Mariri
Author information:
Department of Molecular Biology and Biotechnology, Atomic Energy Commission of Syria, Damascus, Syria
Abstract
Background:
Salmonella enterica serovar Typhimurium is considered one of the most important emerging food-borne pathogens in public health worldwide. Meat is commonly known as the food sources responsible for the salmonellosis outbreak.
Methods: Overall, 141 different meat samples were randomly collected from local markets. The conventional culture method was performed to isolate Salmonella spp., and then, using two pairs of primers, a multiplex polymerase chain reaction (PCR) assay was employed to confirm the identification of isolated colonies as Salmonella spp. and determine serovars as Typhimurium.
Results: Out of 141 samples, 48 (34%) ones were presumptively isolated as Salmonella on the Salmonella agar medium and distributed as 24%, 23%, and 42% among veal, lamb, and chicken meat, respectively. However, the results of multiplex PCR showed that 4.9% of chicken meat was merely identified as S. Typhimurium. In general, S. Typhimurium isolates were found only in chicken meat.
Conclusion: Salmonella Typhimurium isolates were only observed in the chicken meat. Multiplex PCR was found to be a specific and rapid alternative method for the identification of various types of Salmonella.
Keywords: Salmonella Typhimurium, Meat, SA medium, Multiplex PCR
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Safi M, Al Balaa B, Qasem S, Al Hallab L, Al-Mariri A. Isolation and identification of salmonella Typhimurium in retail meat sources by multiplex polymerase chain reaction. Avicenna J Clin Microbiol Infect. 2022; 9(4):152-156. doi:10.34172/ajcmi.2022.3042
Introduction
Typhoid is a serious problem in areas without clean water sources and proper sanitation. It affects around six million people worldwide and is estimated to cause 600 000 deaths a year. The disease is not common in developed countries, and most infections come from abroad or from migrants (1). Approximately 80% of infections and deaths occur in Asia, and the remaining cases belong to Africa and Latin America (2). Salmonella is a bacterium associated with animals. Animal skin and fur infections occur through the transmission of bacteria through the blood from the intestines of infected animals (3). This bacterium causes enteritis in poultry which is associated with a high mortality rate (4). It also causes death in newborn calves (5). These bacteria are not classified as the microbiota of the poultry intestinal flora, but they are introduced from the environment through insects, rodents, and feed. Infected adult animals show no outward signs, and infection can be spread by stable brushes and milking tools (6). Salmonella can also be transmitted between animals or from an infected animal to the veterinarian through the veterinary instruments used for detection (7). Salmonella can be isolated from the liver, spleen, and gallbladder of many species, including mammals (goats, sheep, and horses), birds (pigeons and ducks), Amphibians (frogs), and some fish species such as catfish (8). In 2009, more than 40 000 cases of salmonellosis (13.6 cases per 100 000 people) were tested by public health laboratories in the United States; this represents a decrease of about 15% from the previous year, but an increase of 4.2% from 1996 (9). Overall, the prevalence of Salmonella has not significantly changed since 1996 in the United States (10,11).
Non-typhoid Salmonella is the second most common cause of foodborne illness and causes 35% and 28% of hospital admissions and deaths in the USA, respectively (12). Among the many types of Salmonella, Salmonella Typhimuriumand Salmonella enteritidis are among the most isolated types during foodborne outbreaks in the world (13). S. enteritidis is considered the leading cause of foodborne diseases (14); during recent decades, an increasing number of these bacteria has been isolated around the world (13,15).
Studies using mice play a key role in all areas of medical research. Furthermore, systemic infection patterns in mice have been widely used for more than 20 years. Mice are especially employed in infectious studies because of their anatomical similarity to humans. Mouse salmonellosis is an experimental pattern commonly used to mimic acute infections in humans because the invasive disease caused by S. Typhimurium in mice is similar to the acute phase of human typhoid fever caused by S. typhi (16).
Diseases caused by Salmonella are exhausted to humans and cause the prolonged absence from work. Although mortality is relatively low, the cost of treatment is high, especially as these bacteria are starting to be resistant to many antimicrobial therapy regimens (17,18). It is known that typhi (and paratyphi ) and Typhimurium are the only two types that cause the disease in humans as a result of eating infected meat or drinking contaminated water. Recently, the resistance of these bacteria to antibiotics has increased in our country. Therefore, the current work aimed to isolate and type S. Typhimurium from some Syrian veal, lamb, and chicken meat samples collected from different regions using conventional microbiology detection compared to the ones detected using the multiplex-polymerase chain reaction (PCR) technique. It was attempted to conduct subsequent studies on mice to determine the effectiveness of antibiotics and some alternative therapies against these bacteria.
Materials and Methods
Collection of Samples
One hundred forty-one different meat samples were collected, including 35, 81, and 25 samples of lamb, chicken, and veal meat from different markets in Damascus, respectively.
Isolation of Salmonella
Twenty-five grams of each meat sample were homogenized using a stomacher (Stomacher® 80 Biomaster Bags; Seward Ltd, UK) into 225 mL of buffered peptone water and incubated at 37°C for 16-20 hours. Then, 1 mL was transferred to 10-mL selenite cysteine broth and incubated for 20-24 hours at 37°C (19). The cultivation was performed on the medium of lysine iron agar; it is a differential medium for intestinal bacteria and consists of several materials per liter, including a pancreatic digest of gelatin (5 g), yeast extract (3 g), dextrose (1 g), L-lysine (10 g), ferric ammonium citrate (0.5 g), sodium thiosulfate (0.04 g), bromocresol purple (0.02 g), and agar (13.5 g). The selected isolates were then cultivated on Salmonella agar medium, which is a differential medium for the Salmonella genus. All plates were incubated at 37°C for 24 hours.
Extraction of DNA
The cetyltrimethylammonium bromide method was used to isolate the DNA (20). Briefly, 2 µL of DNA extraction in TE buffer was read using a nanodrop machine. To detect the DNA concentration, TE buffer was employed as blank. Then, the concentration of 100 ng/mL in each sample was made and stored at -20°C.
Multiplex Polymerase Chain Reaction
The multiplex PCR was applied to identify the specific S. Typhimurium isolates. Multiplex PCR amplification was performed using two sets of primer pairs. The primer pair, ST11 (5’-GCCAACCATTGCTAAATTGGCGCA-3’) and ST15 (5’-GGTAGAAATTCCCAGCGGGTACTGG-3’), was specific to Salmonella spp. and targeted a randomly selected sequence of unknown function of 429 bp (21,22). The primer pair, Fli15 (5’-CGGTGTTGCCCAGGTTGGTAAT-3’) and Tym (5’ ACTCTTGCTGGCGGTGCGACTT-3’), was specific to the filiC gene of S. Typhimuriumof 559 bp (19). Multiplex PCR was performed in a final volume of 25 μL containing 200 ng bacterial DNA, 25 pmol of each forward and reversed set primers, 0.2 mM of dNTP Mix, 1.5 mM MgCl2, 1X PCR buffer, and 1U Taq DNA polymerase. The cycling conditions included one cycle at 95°C for 5 minutes, 35 cycles (at 94°C for 60 seconds, at 55°C for 60 seconds, and at 72°C for 60 seconds), then one cycle at 72°C for 10 minutes. The results of multiplex PCR were observed under UV light after electrophoresis in agarose gel pre-stained with ethidium bromide.
Results
Isolation of Salmonella
On lysine iron agar medium, fifty-four positive isolates (38.3%) were obtained, which were distributed as 24%, 25.7%, and 48% among veal, lamb, and chicken meat, respectively, where the medium maintained its violet color, and the colonies appeared transparent with or without dark centers (Figure 1). However, using Salmonella agar medium, forty-eight positive isolates (34%) were obtained, which were distributed as 24%, 22.9%, and 42% among veal, lamb, and chicken meat, respectively; where the medium turned yellow around the isolates, taking a transparent color with or without dark centers (Figure 2). The distributions of Salmonella among different meat sources are shown in Table 1.
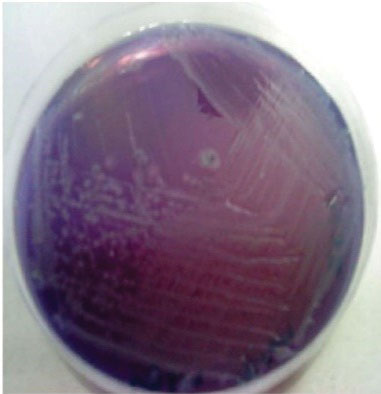
Figure 1.
Appearance of Salmonella Colonies on Lysine Iron Agar Medium. Note. They were locked as transparent colonies with black centers
.
Appearance of Salmonella Colonies on Lysine Iron Agar Medium. Note. They were locked as transparent colonies with black centers

Figure 2.
Appearance of Salmonella Colonies on Salmonella Agar Medium. Note. In this medium, transparent colonies with black spots are observed, and the color of the medium changes to yellow
.
Appearance of Salmonella Colonies on Salmonella Agar Medium. Note. In this medium, transparent colonies with black spots are observed, and the color of the medium changes to yellow
Table 1.
Distributions of Salmonella Among Different Meat Sources: Bacterial Isolation and Multiplex PCR Reaction Results
|
Meat
|
Samples
|
LIA medium
|
SA Medium
|
Multiplex PCR
|
|
Salmonella
spp.
|
S. typhimurium
|
|
No
|
%*
|
No
|
%*
|
No
|
%**
|
No
|
%**
|
| Veal |
25 |
6 |
24 |
6 |
24 |
2 |
8 |
- |
- |
| Lamb |
35 |
9 |
25.7 |
8 |
22.9 |
3 |
8.6 |
- |
- |
| Chicken |
81 |
39 |
48 |
34 |
42 |
29 |
35.8 |
4 |
4.9 |
| Total |
141 |
54 |
38.3 |
48 |
34 |
34 |
24.1 |
4 |
2.9 |
Note. PCR: Polymerase chain reaction; LIA: Lysine iron agar; SA: Salmonella agar. * % compared with total samples; ** % compared with suspected isolation in the SA medium.
Molecular Typing Using Multiplex PCR
The multiplex PCR was performed on all samples, showing positive results on Salmonella agar medium in 6, 8, and 34 samples of veal, lamb, and chicken meat, respectively. The results in Table 1 were obtained after transferring it onto an agarose gel. As a result, only four isolates of S. Typhimurium were obtained in 81 samples of chicken meat (4.9%). However, no isolate of S. Typhimurium was found in veal and lamb meat samples. Table 1 presents the results of cultures and PCR, and Figure 3 shows the data of PCR on the agarose gel.
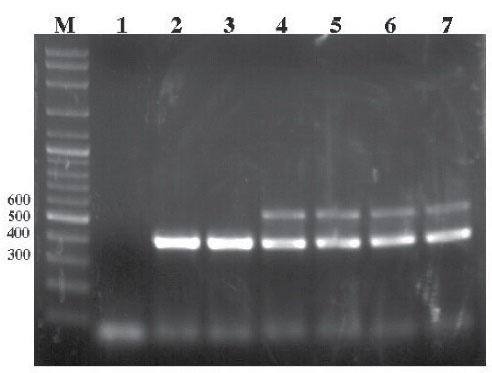
Figure 3.
Agarose Gel Electrophoresis Results by Multiplex PCR. Note. PCR: Polymerase chain reaction; Lane M: GeneRuler DNA ladder (Thermo); Lane 1: Negative control (no template DNA was added); Lanes 2 and 3: Amplification of 429 bp fragment of Salmonella spp.; Lanes 4, 5, 6, and 7: Amplification of 429 and 559 bp fragments of Salmonella Typhimurium.
.
Agarose Gel Electrophoresis Results by Multiplex PCR. Note. PCR: Polymerase chain reaction; Lane M: GeneRuler DNA ladder (Thermo); Lane 1: Negative control (no template DNA was added); Lanes 2 and 3: Amplification of 429 bp fragment of Salmonella spp.; Lanes 4, 5, 6, and 7: Amplification of 429 and 559 bp fragments of Salmonella Typhimurium.
Discussion
In recent decades, Salmonella, especially Typhimurium, has rapidly developed resistance against antibiotics such as ampicillin, chloramphenicol, co-trimoxazole, and even against ciprofloxacin (23,24). In addition, multidrug-resistant enteric fever is a major problem worldwide (25,26).
Animals, particularly poultry, and their products (e.g., eggs and meat) are the main source of human infections caused by this pathogen (27). The epidemiological nature and severe pathogenicity of these pathogens are considered essential agents in food-related diseases. Further, diseases caused by these pathogens are of great importance to public health (28,29).
Almost all types of enteric Salmonella, especially Typhimurium, which infect humans and animals, can infect many types of animals and cause systemic diseases in many animals (30). However, traditional serotyping methods are highly time-consuming and laboratory intensive and require high-quality serological antibodies and extremely well-trained human laboratory staff (31-33).
The typing of Salmonella is essential for monitoring and controlling foodborne diseases. This allows rapid detection, identification of infection sources, and control of pandemics in addition to discovering new serotypes and new transmission routes of infection (34). Because of the great importance of S. Typhimurium as an animal pathogen, researchers have found it more important to reveal its presence than the other bacteria of the Enterobacter genus (35).
Phenotyping methods play an important role in determining the genus of Salmonella. Phenotyping using the Kauffman-White scale is considered in the primary phenotyping of Salmonella and is based on the detection of the presence of somatic and flagellated antigens on the surface of the bacterial cell, while phage profiling is employed to identify subtypes (36,37). However, the presence of some defects associated with serotyping, including cross-sections between serotypes, has encouraged researchers to discover faster and simpler diagnostic methods based on molecular techniques.
To eliminate serotyping-associated problems, PCR and other nucleotide-based methods allowed the acceleration of the detection of serotypes by single gene detection or gene amplification (32).
The economic losses caused by S. Typhimurium in the poultry industry cannot be ignored since this organism plays an important role in foodborne diseases.
Several studies have focused on the isolating and typing of Salmonella spp. and S. Typhimurium. It was found that most cases of Salmonella infection occur through eating contaminated food, especially food of animal origin (38,39).
Salmonella Typhimurium detection has been reported from poultry by several researchers. In a study of 200 samples of field meat from chicken, turkey, and pig, White et al (40) revealed the presence of 8 isolates of S. Typhimurium, four of which were from chicken and four from pigs. While Nagappa et al (41), who used PCR assay, found one S. Typhimurium isolate only in 100 chicken meat samples. In another study, 3% of S. Typhimurium belonged to chicken skin and feathers (42) using multiplex PCR. In contrast, Abd El-Aziz (43) showed that a proportion of 44% of Egyptian chicken meat contains S. Typhimurium.
In our study, only four isolates of S. Typhimurium were observed in 81 chicken meat samples (about 5% of the isolates). This percentage is highly consistent with most studies conducted on chicken meat. However, we observed no S. Typhimurium isolates in the meat of other animals used in our study. This is probably, in one way or another, due to the nature of different feeding of animal species and the contamination of the tools employed in the establishment and breeding of each type of animal.
Conclusions
Salmonella Typhimurium isolates were only detected in chicken meat. Multiplex PCR was found to be a specific, simple, and rapid alternative method for the identification of Salmonella spp. and allowed the observation of specific serovar (Typhimurium) contamination in the field conditions.
Authors’ Contribution
Conceptualization: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri.
Methodology: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri, Samah Qasem, Laila Al Hallab.
Validation: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri.
Formal Analysis: Mazen Safi.
Investigation: Mazen Safi, Bassam Al Balaa.
Resources: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri.
Data Curation: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri.
Writing—Original Draft Preparation: Mazen Safi.
Writing—Review and Editing: Mazen Safi.
Visualization: Mazen Safi, Samah Qasem.
Supervision: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri.
Project Administration: Mazen Safi, Bassam Al Balaa, Ayman Al-Mariri.
Funding Acquisition: Atomic Energy Commission of Syria.
Competing Interests
None to be declared.
Ethical Approval
This article contains no studies with human participants or live animals performed by any of the authors.
References
- Mogasale V, Maskery B, Ochiai RL, Lee JS, Mogasale VV, Ramani E. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health 2014; 2(10):e570-80. doi: 10.1016/s2214-109x(14)70301-8 [Crossref] [ Google Scholar]
- World Health Organization (WHO). The Diagnosis, Treatment and Prevention of Typhoid Fever. WHO/V&B/03.17. Geneva, Switzerland: WHO; 2003.
- Center for Disease Control and Prevention (CDC). Salmonellosis. 2004. http://www.cdc.gov/ncidod/dbmd/diseaseinfo/salmonellosis_g.htm.
- Hamid N, Jain SK. Immunological, cellular and molecular events in typhoid fever. Indian J Biochem Biophys 2007; 44(5):320-30. [ Google Scholar]
- McKinley Health Center (MHC). Salmonella. 2004. www.mckinley.uiuc.edu.
- Lavigne JP, Blanc-Potard AB. Molecular evolution of Salmonella enterica serovar Typhimurium and pathogenic Escherichia coli: from pathogenesis to therapeutics. Infect Genet Evol 2008; 8(2):217-26. doi: 10.1016/j.meegid.2007.11.005 [Crossref] [ Google Scholar]
- Ly KT, Casanova JE. Mechanisms of Salmonella entry into host cells. Cell Microbiol 2007; 9(9):2103-11. doi: 10.1111/j.1462-5822.2007.00992.x [Crossref] [ Google Scholar]
- Hoelzer K, Moreno Switt AI, Wiedmann M. Animal contact as a source of human non-typhoidal salmonellosis. Vet Res 2011; 42(1):34. doi: 10.1186/1297-9716-42-34 [Crossref] [ Google Scholar]
- Center for Disease Control and Prevention (CDC). Salmonella Annual Summary Tables 2009. http://www.cdc.gov/ncezid/dfwed/PDFs/SalmonellaAnnualSummaryTables2009.pdf.
- Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis 2004; 38 Suppl 3:S127-34. doi: 10.1086/381578 [Crossref] [ Google Scholar]
- Miller S, Pegues D. Salmonella species, including Salmonella typhi. In: Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Elsevier; 2015.
- Center for Disease Control and Prevention (CDC). Estimates of Foodborne Illness in the United States. 2011. http://www.cdc.gov/foodborneburden/2011-foodborneestimates.html.
- Herikstad H, Motarjemi Y, Tauxe RV. Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect 2002; 129(1):1-8. doi: 10.1017/s0950268802006842 [Crossref] [ Google Scholar]
- Brenner FW, Villar RG, Angulo FJ, Tauxe R, Swaminathan B. Salmonella nomenclature. J Clin Microbiol 2000; 38(7):2465-7. doi: 10.1128/jcm.38.7.2465-2467.2000 [Crossref] [ Google Scholar]
- Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev 2015; 28(4):901-37. doi: 10.1128/cmr.00002-15 [Crossref] [ Google Scholar]
- Mathur R, Oh H, Zhang D, Park SG, Seo J, Koblansky A. A mouse model of Salmonella typhi infection. Cell 2012; 151(3):590-602. doi: 10.1016/j.cell.2012.08.042 [Crossref] [ Google Scholar]
- Firoozeh F, Zahraei-Salehi T, Shahcheraghi F, Karimi V, Aslani MM. Characterization of class I integrons among Salmonella enterica serovar Enteritidis isolated from humans and poultry. FEMS Immunol Med Microbiol 2012; 64(2):237-43. doi: 10.1111/j.1574-695X.2011.00883.x [Crossref] [ Google Scholar]
- Ranjbar R, Rahmati H, Shokoohizadeh L. Detection of common clones of Salmonella enterica serotype Infantis from human sources in Tehran hospitals. Gastroenterol Hepatol Bed Bench 2018; 11(1):54-9. [ Google Scholar]
- Nordic Method Committee on Food Analysis. NMKL method no 71, Salmonella. Detection in food. Åbo, Finland; 1999.
- Ali R, Al-Achkar K, Al-Mariri A, Safi M. Role of polymerase chain reaction (PCR) in the detection of antibiotic-resistant Staphylococcus aureus. Egypt J Med Hum Genet 2014; 15(3):293-8. doi: 10.1016/j.ejmhg.2014.05.003 [Crossref] [ Google Scholar]
- Aabo S, Rasmussen OF, Roseen L, Sørensen PD, Olsen JE. Salmonella identification by the polymerase chain reaction. Mol Cell Probes 1993; 7(3):171-8. doi: 10.1006/mcpr.1993.1026 [Crossref] [ Google Scholar]
- Soumet C, Ermel G, Rose V, Rose N, Drouin P, Salvat G. Identification by a multiplex PCR-based assay of Salmonella Typhimurium and Salmonella Enteritidis strains from environmental swabs of poultry houses. Lett Appl Microbiol 1999; 29(1):1-6. doi: 10.1046/j.1365-2672.1999.00559.x [Crossref] [ Google Scholar]
- Mandal S, Mandal MD, Pal NK. Antimicrobial resistance pattern of Salmonella typhi isolates in Kolkata, India during 1991-2001: a retrospective study. Jpn J Infect Dis 2002; 55(2):58-9. [ Google Scholar]
- Pal NK, Mandal S, Mandal MD. Scattergram analysis to explore the emerging problem related to in vitro susceptibility test for Salmonella enterica serovar Typhi to ciprofloxacin. Int J Antimicrob Agents 2004; 24(3):297-9. doi: 10.1016/j.ijantimicag.2004.02.025 [Crossref] [ Google Scholar]
- Cuypers WL, Jacobs J, Wong V, Klemm EJ, Deborggraeve S, Van Puyvelde S. Fluoroquinolone resistance in Salmonella: insights by whole-genome sequencing. Microb Genom 2018; 4(7):e000195. doi: 10.1099/mgen.0.000195 [Crossref] [ Google Scholar]
- Threlfall EJ, Day M, de Pinna E, Lewis H, Lawrence J. Drug-resistant enteric fever in the UK. Lancet 2006; 367(9522):1576. doi: 10.1016/s0140-6736(06)68691-1 [Crossref] [ Google Scholar]
- Mahé A, Bougeard S, Huneau-Salaün A, Le Bouquin S, Petetin I, Rouxel S. Bayesian estimation of flock-level sensitivity of detection of Salmonella spp, Enteritidis and Typhimurium according to the sampling procedure in French laying-hen houses. Prev Vet Med 2008; 84(1-2):11-26. doi: 10.1016/j.prevetmed.2007.10.003 [Crossref] [ Google Scholar]
- Kottwitz LB, Back A, Leão JA, Alcocer I, Karan M, Oliveira TC. Salmonella contamination in an egg production chain of a laying hens integration. Arq Bras Med Vet Zootec 2008; 60(2):496-8. doi: 10.1590/s0102-09352008000200034 [Crossref] [ Google Scholar]
- Aarestrup FM, Hendriksen RS, Lockett J, Gay K, Teates K, McDermott PF. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg Infect Dis 2007; 13(5):726-31. doi: 10.3201/eid1305.061489 [Crossref] [ Google Scholar]
- Heithoff DM, Shimp WR, Lau PW, Badie G, Enioutina EY, Daynes RA. Human Salmonella clinical isolates distinct from those of animal origin. Appl Environ Microbiol 2008; 74(6):1757-66. doi: 10.1128/aem.02740-07 [Crossref] [ Google Scholar]
- Rementeria A, Vivanco AB, Ramirez A, Hernando FL, Bikandi J, Herrera-León S. Characterization of a monoclonal antibody directed against Salmonella enterica serovar Typhimurium and serovar [4,5,12:i:-]. Appl Environ Microbiol 2009; 75(5):1345-54. doi: 10.1128/aem.01597-08 [Crossref] [ Google Scholar]
- Hong Y, Liu T, Lee MD, Hofacre CL, Maier M, White DG. Rapid screening of Salmonella enterica serovars Enteritidis, Hadar, Heidelberg and Typhimurium using a serologically-correlative allelotyping PCR targeting the O and H antigen alleles. BMC Microbiol 2008; 8:178. doi: 10.1186/1471-2180-8-178 [Crossref] [ Google Scholar]
- Gallegos-Robles MA, Morales-Loredo A, Alvarez-Ojeda G, Vega PA, Chew MY, Velarde S. Identification of Salmonella serotypes isolated from cantaloupe and chile pepper production systems in Mexico by PCR-restriction fragment length polymorphism. J Food Prot 2008; 71(11):2217-22. doi: 10.4315/0362-028x-71.11.2217 [Crossref] [ Google Scholar]
- Fitzgerald C, Sherwood R, Gheesling LL, Brenner FW, Fields PI. Molecular analysis of the rfb O antigen gene cluster of Salmonella enterica serogroup O:6,14 and development of a serogroup-specific PCR assay. Appl Environ Microbiol 2003; 69(10):6099-105. doi: 10.1128/aem.69.10.6099-6105.2003 [Crossref] [ Google Scholar]
- Lim YH, Hirose K, Izumiya H, Arakawa E, Takahashi H, Terajima J. Multiplex polymerase chain reaction assay for selective detection of Salmonella enterica serovar Typhimurium. Jpn J Infect Dis 2003; 56(4):151-5. [ Google Scholar]
- Bale JA, de Pinna EM, Threlfall E, Ward LR. Kauffmann-White Scheme-2007: Salmonella Identification: Serotypes and Antigen Formulae. London: Centre for Infections, Health Protection Agency; 2007.
- Nair S, Lin TK, Pang T, Altwegg M. Characterization of Salmonella serovars by PCR-single-strand conformation polymorphism analysis. J Clin Microbiol 2002; 40(7):2346-51. doi: 10.1128/jcm.40.7.2346-2351.2002 [Crossref] [ Google Scholar]
- Buzby JC, Roberts T. The economics of enteric infections: human foodborne disease costs. Gastroenterology 2009; 136(6):1851-62. doi: 10.1053/j.gastro.2009.01.074 [Crossref] [ Google Scholar]
- Ranjbar R, Elhaghi P, Shokoohizadeh L. Multilocus sequence typing of the clinical isolates of Salmonella enterica serovar Typhimurium in Tehran hospitals. Iran J Med Sci 2017; 42(5):443-8. [ Google Scholar]
- White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S. The isolation of antibiotic-resistant Salmonella from retail ground meats. N Engl J Med 2001; 345(16):1147-54. doi: 10.1056/NEJMoa010315 [Crossref] [ Google Scholar]
- Nagappa K, Tamuly S, Saxena MK, Singh SP. Isolation of Salmonella Typhimurium from poultry eggs and meat of Tarai region of Uttaranchal. Indian J Biotechnol 2007; 6(3):407-9. [ Google Scholar]
- Paião FG, Arisitides LG, Murate LS, Vilas-Bôas GT, Vilas-Boas LA, Shimokomaki M. Detection of Salmonella spp, Salmonella Enteritidis and Typhimurium in naturally infected broiler chickens by a multiplex PCR-based assay. Braz J Microbiol 2013; 44(1):37-41. doi: 10.1590/s1517-83822013005000002 [Crossref] [ Google Scholar]
- El-Aziz DM. Detection of Salmonella typhimurium in retail chicken meat and chicken giblets. Asian Pac J Trop Biomed 2013; 3(9):678-81. doi: 10.1016/s2221-1691(13)60138-0 [Crossref] [ Google Scholar]