Avicenna Journal of Clinical Microbiology and Infection. 9(4):165-170.
doi: 10.34172/ajcmi.2022.3395
Original Article
Molecular Characterization and Antibiotic Resistance Profile of Methicillin-Resistant Staphylococcus aureus (MRSA) Strains Isolated From Milk Samples of Apparently Healthy Cattle in Hamedan, Iran
Hossein Ghaderi 1  , Abdolmajid Mohammadzadeh 1, *
, Abdolmajid Mohammadzadeh 1, *  , Mohamadreza Pajohi-alamoti 2, Ali Sadeghi-nasab 3, Pezhman Mahmoodi 1, Ali Goudarztalejerdi 1
, Mohamadreza Pajohi-alamoti 2, Ali Sadeghi-nasab 3, Pezhman Mahmoodi 1, Ali Goudarztalejerdi 1 
Author information:
1Department of Pathobiology, Faculty of Veterinary Science, Bu-Ali Sina University, Hamedan, Iran
2Department of Food Hygiene and Quality Control, Faculty of Veterinary Science, Bu-Ali Sina University, Hamedan, Iran
3Department of Clinical Sciences, Faculty of Veterinary Science, Bu-Ali Sina University, Hamedan, Iran
Abstract
Background: Staphylococcus aureus, as a major food-borne pathogen, is the most commonly isolated bacterium from bovine mastitis. However, some S. aureus strains exhibit a high rate of antibiotic resistance, among which, methicillin-resistant S. aureus (MRSA) is very important. The present study was conducted to isolate, characterize, and determine the antibiotic resistance profile of MRSA strains in milk.
Methods: Staphylococcus aureus strains were isolated and identified from 415 milk samples collected from apparently healthy cattle in Hamedan province, Iran. Molecular characteristics of the strains were identified using multiplex polymerase chain reaction (PCR) and the antibiotic resistance profile of the isolates was determined by Kirby-Bauer disk diffusion susceptibility test.
Results: A total of 76 S. aureus strains were isolated and identified. The PCR results indicated that 50 (65.78%) isolates possessed mecA gene and were found to be MRSA strains. Twelve isolates (15.78%) showed phenotypic resistance to oxacillin in disk diffusion method. All 76 S. aureus isolates (100%) were resistant to penicillin and susceptible to ciprofloxacin and gentamicin.
Conclusion: The results of the present study indicated that bovine milk may contain MRSA strains and this is worrying as these isolates may transfer multi-drug resistance to the isolates that circulate among humans, animals, and food chains.
Keywords: Bovine mastitis, Milk, Methicillin-resistant Staphylococcus aureus, Multi-drug resistance
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Ghaderi H, Mohammadzadeh A, Pajohi-alamoti M, Sadeghi-nasab A, Mahmoodi P, Goudarztalejerdi A. Molecular characterization and antibiotic resistance profile of methicillin-resistant staphylococcus aureus (mrsa) strains isolated from milk samples of apparently healthy cattle in hamedan, iran. Avicenna J Clin Microbiol Infect. 2022; 9(4):165-170. doi:10.34172/ajcmi.2022.3395
Introduction
Staphylococcus aureus is an opportunistic bacterial pathogen which causes a wide range of illnesses such as impetigo, cellulitis, pneumonia, osteomyelitis, endocarditis, bacteremia, and toxic shock syndrome and food poisoning in humans, as well as mastitis, dermatitis, and arthritis in animals worldwide (1-3). Among these diseases, bovine mastitis has been known as the most prevalent and costly disease in dairy industry worldwide (4,5).
Various microorganisms including bacteria, viruses, fungi, and algae are involved in bovine mastitis, among which staphylococci, streptococci, and Gram-negative bacilli are the major etiological agents of bovine mastitis (6-9). Staphylococcus aureus is the most common causative agent and frequently isolated bacterium from bovine mastitis (6,10-13). In Iran, various investigations based on conventional microbiology methods and molecular techniques described some characteristics and antibiotic susceptibility profile of S. aureus strains as a major causative agent of bovine mastitis (14,15). However, increased resistance of S. aureus,isolated from mastitic cows to a wide array of commonly used antibiotics, has been reported in several studies (16-18).
Different groups of antibiotics including β-lactams, tetracycline, macrolides, and so on are commonly used to treat bovine mastitis. Hence, it is important to monitor antibiotic resistance patterns of S. aureus isolates, especially those which are resistant to methicillin. Methicillin is one of the recently developed antibiotics, and resistance against this antibiotic is very important. S. aureus isolates are divided into two groups: methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus (MSSA). Consequently, the presence of MRSA strains in milk samples could be a major concern for public health.
Resistance to methicillin in S. aureus is mediated by mecA gene which is located on the chromosome and encodes a penicillin-binding protein 2a with reduced affinity for β-lactams (19,20). Therefore, mecA gene can be used as a molecular marker to detect methicillin-resistant isolates. However, other factors such as femA operon, which contains regulatory genes, are essential for the expression of methicillin-resistance in S. aureus as well (21). However, fem genes were suggested to be specific for S. aureus and can be used for molecular identification of S. aureus isolates (22).
Given that the presence of MRSA strains, as major food-borne bacteria, in a common food source like milk is very important in terms of public health and the spread of antibiotic resistance in bacterial populations, the present study was conducted to isolate, characterize, and determine antibiotic resistance profile of MRSA strains in cattle withno clinicalsignsin dairy farms of Hamedan province, west of Iran.
Materials and Methods
Sampling
A total of 415 milk samples were collected from healthy cattle (the cattle with a good appetite, normal rumination, normal milk production, apparently healthy milk, normal udder tissue, and with no clinical signs of mastitis) belonging to 7 dairy farms in Hamedan province of Iran. These milk samples were collected in two seasons, warm-dry (n = 234) and cold-wet (n = 181). Microscopic somatic cell count (SCC) and California mastitis test (CMT) were carried out on the collected milk samples for direct and indirect estimation of the number of somatic cells, respectively. SCC and CMT are known as reliable indicators of chronic intramammary infection (23). Milk samples which contained over 300 000 cells/mL were considered positive cases of subclinical/chronic bovine mastitis in SCC test, while a score of one or more was considered positive in CMT (24).
Isolation and Identification of Staphylococcus aureus
Milk samples (100 µL) were inoculated on 5% sheep blood agar (Merck, Germany) at 37ºC for 24 hours. Afterwards, conventional microbiological tests including microscopic examination, catalase test, coagulase test, and attributes on the mannitol salt agar and DNase agar (Merck, Germany) were carried out on the suspected colonies for the identification of S. aureus.
Antimicrobial Susceptibility of Staphylococcus aureus Isolates
Staphylococcus aureus isolates were tested for antimicrobial susceptibility to 9 antibiotics: penicillin (16 IU), ciprofloxacin (5 µg), oxacillin (1 µg), chloramphenicol (30 µg), gentamicin (10 µg), vancomycin (30 µg), streptomycin (10 µg), tetracycline (30 µg), and cefixime (5 µg). All antibiotic discs were purchased from Padtan Teb® (Padtan Teb Co, Iran). Antimicrobial susceptibility testing was performed on Mueller-Hinton agar (Merck, Germany) using disc diffusion method, as described in the guidelines of the Clinical and Laboratory Standards Institute (CLSI 2019). The results were categorized as susceptible, intermediate, or resistant according to the above-mentioned guidelines (25). Quality control was performed using S. aureus ATCC 25923 as the reference strain.
DNA Extraction
Total DNA was extracted from the phenotypically characterized bacterial isolates using the previously described method with some modifications. Briefly, the isolates were grown in nutrient broth at 37°C for 24 hours. Thereafter, 3 mL of bacterial suspension was centrifuged at 8000 rpm for 3 minutes and 200 μL of a lysis buffer (1% Triton X-100, 0.5% Tween 20, 10 mM Tris-HCl, 1 mM EDTA, pH = 8.0) was added to the pellets and microtubes were incubated in a boiling water bath (100°C) for 10 minutes, followed by centrifugation at 10 000 rpm for 2 minutes. The supernatants were transferred into clean microtubes, and 3-5 μL of each sample was used as template DNA in polymerase chain reaction (PCR) assays (26).
Multiplex Polymerase Chain Reaction
Staphylococcus aureus isolates were examined by a multiplex PCR assay which simultaneously targeted both femA, a species-specific gene for definite identification of S. aureus, and mecA genes to genetically detect MRSA strains using primers previously described (27). The characteristics of the primers are given in Table 1.
Table 1.
Oligonucleotide Primers Used in the PCR Assay
|
Primer
|
Oligonucleotide Sequence (5'- 3')
|
Target Gene
|
Amplicon Size (bp)
|
GFEMAR-1
GFEMAR-2 |
AAAAAAGCACATAACAAGCG
GATAAAGAAGAAACCAGCAG |
femA
|
132 |
GMECAR-1
GMECAR-2 |
ACTGCTATCCACCCTCAAAC
CTGGTGAAGTTGTAATCTGG |
mecA
|
163 |
PCR amplifications were performed in a SimpliAmpTM Thermal Cycler (Applied Biosystems, USA). The reaction mixtures consisted of 3-5 µL of DNA template, 12.5 μL of 2X Master Mix (Ampliqon, Denmark), and 1 µL of each primer pair (25 pmol- TakapouZist, Iran), and the final volume of the reaction mixture was brought up to 25 µL using distilled deionized water. PCR program for amplification of femA and mecA genes consisted of initial denaturation at 94ºC for 5 minutes, 35 cycles of amplification with denaturation at 94ºC for 2 minutes, annealing at 57ºC for 2 minutes, extension at 72ºC for 1 minute, and a final extension at 72ºC for 1 minute. The PCR products were analyzed in 2% agarose gel containing 0.5 µg/mL of ethidium bromide and subjected to electrophoresis in 1X TAE buffer. Gels were visualized under UV light and documented using UVITEC gel documentation system (Transilluminator, France). A sample containing no DNA was used as the negative control in all PCR runs. Furthermore, standard methicillin-susceptible S. aureus (MSSA, ATCC 25 923) and methicillin-resistant S. aureus (MRSA, ATCC 33 591) were used as controls for PCR optimization (28).
Statistical Analysis
The data were analyzed using student’s t test by SAS software volume 8.2, and P < 0.05 was considered to be statistically significant.
Results
In this study, the results of SCC test and CMT on the 415 collected milk samples showed that 114 (27.47%) and 132 (31.81%) samples were positive in CMT and SCC test, respectively. There was no statistically significant difference between the results of CMT and SCC test for the collected milk samples in the two seasons (P > 0.05). However, the correlation between the results of the two methods was high (r = 0.948, P < 0.05).
Staphylococcus aureus was isolated from 83 out of 415 milk samples. Seventy-six isolates (18.31%) showed the expected DNA fragment of 132 bp in PCR and were genotypically identified as S. aureus (Figure 1). In warm-dry and cold-wet seasons, 51 and 25 S. aureus isolateswereidentified by genotypic method, respectively.
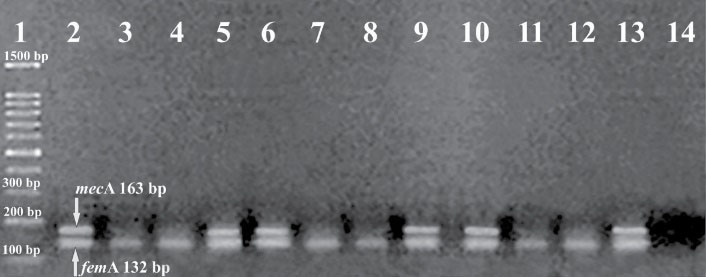
Figure 1.
Agarose Gel Electrophoresis of Multiplex PCR Products of femA and mecA Genes in Staphylococcus aureus. Lane 1: a 100-bp ladder, Lane 2: S. aureus ATCC 33591, positive control for femAand mecA genes. Lanes 3-13: femA -positive S. aureus isolates. Lanes 5, 6, 9, 10 and 13: mecA-positive S. aureus isolates. Lane 14: negative control (contained no template)
.
Agarose Gel Electrophoresis of Multiplex PCR Products of femA and mecA Genes in Staphylococcus aureus. Lane 1: a 100-bp ladder, Lane 2: S. aureus ATCC 33591, positive control for femAand mecA genes. Lanes 3-13: femA -positive S. aureus isolates. Lanes 5, 6, 9, 10 and 13: mecA-positive S. aureus isolates. Lane 14: negative control (contained no template)
The mecA gene was detected in 50 (65.78%) out of 76 isolated S. aureus (Figure 1), among which 12 (24%) isolates phenotypically showed resistance to oxacillin.
Meanwhile, the antimicrobial susceptibility test (disc diffusion method) was done to check the resistance of the isolates to various antibiotics including penicillin, oxacillin, ciprofloxacin, tetracycline, gentamicin, streptomycin, vancomycin, chloramphenicol, and cefixime and the results indicated that all of the 76 S. aureus isolates (100%) were resistant to penicillin and susceptible to ciprofloxacin and gentamicin. The detailed resistance profiles of the isolates are presented in Table 2.
Table 2.
The Antibiotic Resistance Profile of the Staphylococcus aureus Isolates
|
Antibiotic Agent
|
Resistant No. (%)
|
Intermediate No. (%)
|
Susceptible No. (%)
|
| Penicillin |
76 (100) |
0 (0) |
0 (0) |
| Ciprofloxacin |
0 (0) |
0 (0) |
76 (100) |
| Oxacillin |
12 (15.78) |
0 (0) |
64 (84.21) |
| Chloramphenicol |
1 (1.31) |
73 (96.05) |
2 (2.63) |
| Gentamicin |
0 (0) |
0 (0) |
76 (100) |
| Vancomycin |
13 (17.1) |
0 (0) |
63 (82.89) |
| Streptomycin |
28 (36.84) |
11(14.47) |
39 (51.31) |
| Tetracycline |
30 (39.47) |
2 (2.63) |
44 (57.89) |
| Cefixime |
9 (11.84) |
8 (10.52) |
59 (77.63) |
Discussion
Bovine mastitis is one of the most important diseases affecting the dairy industry worldwide, and S. aureus is the main causative agent of bovine mastitis which responds poorly to antimicrobial therapy (29,30). The prevalence of S. aureus in our study was similar to previous reports from Iran and other parts of the world. Hashemi et al reported that the most frequently isolated bacteria as the cause of mastitis were coagulase-positive staphylococci in Fars province, south of Iran (31). Besides, S. aureus prevalence rates of 10 to 30% have been documented in mastitis-affected cattle (7). Makovec and Ruegg reported that S. aureus was isolated from 9.7% of cows in the United States (32).
In this study, 76 isolates were confirmed as S. aureus using PCR technique. Several studies suggested molecular technique as a specific, highly sensitive, and rapid method for detection and identification of S. aureus (33-35).
The current study results revealed that in the warm-dry season, the prevalence of S. aureus was higher compared to the cold-wet season, which is in accordance with the results of a study conducted by Koivula et al, indicating that S. aureus was more prevalent in spring and November in Southern Finland (36).
The results of this study indicated that S. aureus caused an increase of the SCC in the collected milk samples, which is in agreement with the findings of Jánosi and Baltay (23) and Balemi et al (37). They reported that all pathogens (including S. aureus) resulted in a significant increase of the SCC in individual bulk milk samples collected from dairy cows, camels, and goats.
The current study results indicated that 50 out of 76 isolates (65.78%) possessed mecA gene and were found to be MRSA strains. The present findings about mecA gene were in accordance with other studies which reported that molecular techniques such as multiplex PCR can be of great use for the diagnosis of methicillin-resistant bacteria (7,16,22,38). However, some researchers described that the mecA gene is carried by 95% of S. aureus isolates that display a phenotype of methicillin resistance (39). Results of a study performed by Pérez-Rothet al on various S. aureus strains isolated from patients in Spain revealed that mecA gene was present in 29 (58%) isolates (40).
In this study, the frequency of antibiotic resistance in the tested S. aureus strains was similar to that found in another study in Iran (41-43). Previous studies examining the antimicrobial susceptibility of S. aureus isolated from bovine mastitis demonstrated a high frequency of resistance to tetracycline, penicillin, and erythromycin among S. aureus isolates (12,16,44,45). In this study, gentamicin and ciprofloxacin were completely effective against S. aureus and 100% of the isolates were susceptible to the mentioned drugs. This result was in agreement with reports which indicated that gentamicin was very active against S. aureus (12).
Moreover, of 50 isolates containing mecA gene, 12 (24%) isolates phenotypically showed resistance to oxacillin. Khazaie and Ahmadi reported that among the 95 isolates of S. aureus from bovine subclinical mastitis in Iran, 11 (11.57%) strains were recognized as MRSA (41). In addition, various studies reported that there are some strains which are phenotypically resistant to oxacillin despite the fact that they carry mecA gene (46-48). These studies support our findings.
Conclusion
The present study was a survey on the molecular characterizations and antibiotic resistance profile of S. aureus isolated from milk samples of cattle with no clinical signs in Hamedan, north-west of Iran. The strains had a high frequency of resistance to penicillin indicating that this antibiotic is not suitable for use in dairy cattle herds in this region. However, the recommended antibiotics for the treatment of mastitis are gentamicin and ciprofloxacin. Besides, a large number of MRSA strains were identified in the present study which is absolutely a major concern for public health. Given that even apparently healthy milk may contain MRSA strains and as these isolates can circulate among humans, animals, and food chains, appropriate hygiene measures should be taken to control and decrease infections caused by these bacteria. Meanwhile, it is strongly recommended that antibiotic susceptibility of isolates should be checked before any treatment.
Acknowledgments
The authors are grateful for financial support by research grants from Bu-Ali Sina University of Hamedan.
Authors’ Contribution
Conceptualization: Abdolmajid Mohammadzadeh, Mohamadreza Pajohi-Alamoti, and Ali Sadeghi-Nasab.
Funding acquisition: Abdolmajid Mohammadzadeh, Mohamadreza Pajohi-Alamoti.
Methodology: Hossein Ghaderi, AM, and Ali Sadeghi-Nasab.
Project administration: Abdolmajid Mohammadzadeh, Mohamadreza Pajohi-Alamoti.
Supervision: Abdolmajid Mohammadzadeh, Mohamadreza Pajohi-Alamoti.
Writing – original draft: Ali Goudarztalejerdi.
Writing – review & editing: Ali Goudarztalejerdi, Abdolmajid Mohammadzadeh, and Pezhman Mahmoodi.
All authors have read and approved the final version of the manuscript.
Competing Interests
The authors declare that there is no conflict of interest.
Ethical Approval
All samples were collected from industrial dairy farms and sent to the laboratory under supervision of a veterinarian.
References
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 2015; 28(3):603-61. doi: 10.1128/cmr.00134-14 [Crossref] [ Google Scholar]
- Lozano C, Gharsa H, Ben Slama K, Zarazaga M, Torres C. Staphylococcus aureus in animals and food: methicillin resistance, prevalence and population structure a review in the African continent. Microorganisms 2016; 4(1):12. doi: 10.3390/microorganisms4010012 [Crossref] [ Google Scholar]
- Youssef CRB, Kadry AA, El-Ganiny AM. Investigating the relation between resistance pattern and type of staphylococcal cassette chromosome mec (SCCmec) in methicillin-resistant Staphylococcus aureus. Iran J Microbiol 2022; 14(1):56-66. doi: 10.18502/ijm.v14i1.8802 [Crossref] [ Google Scholar]
- Nunes SF, Bexiga R, Cavaco LM, Vilela CL. Technical note: Antimicrobial susceptibility of Portuguese isolates of Staphylococcus aureus and Staphylococcus epidermidis in subclinical bovine mastitis. J Dairy Sci 2007; 90(7):3242-6. doi: 10.3168/jds.2006-739 [Crossref] [ Google Scholar]
- Raspanti CG, Bonetto CC, Vissio C, Pellegrino MS, Reinoso EB, Dieser SA. Prevalence and antibiotic susceptibility of coagulase-negative Staphylococcus species from bovine subclinical mastitis in dairy herds in the central region of Argentina. Rev Argent Microbiol 2016; 48(1):50-6. doi: 10.1016/j.ram.2015.12.001 [Crossref] [ Google Scholar]
- De Oliveira AP, Watts JL, Salmon SA, Aarestrup FM. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and the United States. J Dairy Sci 2000; 83(4):855-62. doi: 10.3168/jds.S0022-0302(00)74949-6 [Crossref] [ Google Scholar]
- Jagielski T, Puacz E, Lisowski A, Siedlecki P, Dudziak W, Międzobrodzki J. Short communication: antimicrobial susceptibility profiling and genotyping of Staphylococcus aureus isolates from bovine mastitis in Poland. J Dairy Sci 2014; 97(10):6122-8. doi: 10.3168/jds.2014-8321 [Crossref] [ Google Scholar]
- Persson Y, Nyman AK, Grönlund-Andersson U. Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Vet Scand 2011; 53(1):36. doi: 10.1186/1751-0147-53-36 [Crossref] [ Google Scholar]
- Tenhagen BA, Köster G, Wallmann J, Heuwieser W. Prevalence of mastitis pathogens and their resistance against antimicrobial agents in dairy cows in Brandenburg, Germany. J Dairy Sci 2006; 89(7):2542-51. doi: 10.3168/jds.S0022-0302(06)72330-X [Crossref] [ Google Scholar]
- Taponen S, Pyörälä S. Coagulase-negative staphylococci as cause of bovine mastitis- not so different from Staphylococcus aureus?. Vet Microbiol 2009; 134(1-2):29-36. doi: 10.1016/j.vetmic.2008.09.011 [Crossref] [ Google Scholar]
- Barkema HW, Schukken YH, Zadoks RN. Invited review: the role of cow, pathogen, and treatment regimen in the therapeutic success of bovine Staphylococcus aureus mastitis. J Dairy Sci 2006; 89(6):1877-95. doi: 10.3168/jds.S0022-0302(06)72256-1 [Crossref] [ Google Scholar]
- Gentilini E, Denamiel G, Llorente P, Godaly S, Rebuelto M, DeGregorio O. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Argentina. J Dairy Sci 2000; 83(6):1224-7. doi: 10.3168/jds.S0022-0302(00)74988-5 [Crossref] [ Google Scholar]
- Botrel MA, Haenni M, Morignat E, Sulpice P, Madec JY, Calavas D. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhône-Alpes, France. Foodborne Pathog Dis 2010; 7(5):479-87. doi: 10.1089/fpd.2009.0425 [Crossref] [ Google Scholar]
- Hakimi Alni R, Mohammadzadeh A, Mahmoodi P. Molecular typing of Staphylococcus aureus of different origins based on the polymorphism of the spa gene: characterization of a novel spa type. 3 Biotech 2018; 8(1):58. doi: 10.1007/s13205-017-1061-6 [Crossref] [ Google Scholar]
- Sahebekhtiari N, Nochi Z, Eslampour MA, Dabiri H, Bolfion M, Taherikalani M. Characterization of Staphylococcus aureus strains isolated from raw milk of bovine subclinical mastitis in Tehran and Mashhad. Acta Microbiol Immunol Hung 2011; 58(2):113-21. doi: 10.1556/AMicr.58.2011.2.4 [Crossref] [ Google Scholar]
- Gao J, Ferreri M, Yu F, Liu X, Chen L, Su J. Molecular types and antibiotic resistance of Staphylococcus aureus isolates from bovine mastitis in a single herd in China. Vet J 2012; 192(3):550-2. doi: 10.1016/j.tvjl.2011.08.030 [Crossref] [ Google Scholar]
- Sakwinska O, Morisset D, Madec JY, Waldvogel A, Moreillon P, Haenni M. Link between genotype and antimicrobial resistance in bovine mastitis-related Staphylococcus aureus strains, determined by comparing Swiss and French isolates from the Rhône Valley. Appl Environ Microbiol 2011; 77(10):3428-32. doi: 10.1128/aem.02468-10 [Crossref] [ Google Scholar]
- Pitkälä A, Haveri M, Pyörälä S, Myllys V, Honkanen-Buzalski T. Bovine mastitis in Finland 2001--prevalence, distribution of bacteria, and antimicrobial resistance. J Dairy Sci 2004; 87(8):2433-41. doi: 10.3168/jds.S0022-0302(04)73366-4 [Crossref] [ Google Scholar]
- Normanno G, Firinu A, Virgilio S, Mula G, Dambrosio A, Poggiu A. Coagulase-positive staphylococci and Staphylococcus aureus in food products marketed in Italy. Int J Food Microbiol 2005; 98(1):73-9. doi: 10.1016/j.ijfoodmicro.2004.05.008 [Crossref] [ Google Scholar]
- Haenni M, Galofaro L, Ythier M, Giddey M, Majcherczyk P, Moreillon P. Penicillin-binding protein gene alterations in Streptococcus uberis isolates presenting decreased susceptibility to penicillin. Antimicrob Agents Chemother 2010; 54(3):1140-5. doi: 10.1128/aac.00915-09 [Crossref] [ Google Scholar]
- Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol 1995; 33(11):2864-7. doi: 10.1128/jcm.33.11.2864-2867.1995 [Crossref] [ Google Scholar]
- Kobayashi N, Wu H, Kojima K, Taniguchi K, Urasawa S, Uehara N. Detection of mecA, femA, and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect 1994; 113(2):259-66. doi: 10.1017/s0950268800051682 [Crossref] [ Google Scholar]
- Jánosi S, Baltay Z. Correlations among the somatic cell count of individual bulk milk, result of the California Mastitis Test and bacteriological status of the udder in dairy cows. Acta Vet Hung 2004; 52(2):173-83. doi: 10.1556/AVet.52.2004.2.6 [Crossref] [ Google Scholar]
- Dohoo IR, Meek AH. Somatic cell counts in bovine milk. Can Vet J 1982; 23(4):119-25. [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 29th ed. CLSI supplement M100. Wayne, PA: CLSI; 2019.
- Reischl U, Linde HJ, Metz M, Leppmeier B, Lehn N. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence PCR. J Clin Microbiol 2000; 38(6):2429-33. doi: 10.1128/jcm.38.6.2429-2433.2000 [Crossref] [ Google Scholar]
- Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol 2000; 38(3):1032-5. doi: 10.1128/jcm.38.3.1032-1035.2000 [Crossref] [ Google Scholar]
- Kim SY, Kim J, Jeong SI, Jahng KY, Yu KY. Antimicrobial effects and resistant regulation of magnolol and honokiol on methicillin-resistant Staphylococcus aureus. Biomed Res Int 2015; 2015:283630. doi: 10.1155/2015/283630 [Crossref] [ Google Scholar]
- Molineri AI, Camussone C, Zbrun MV, Suárez Archilla G, Cristiani M, Neder V. Antimicrobial resistance of Staphylococcus aureus isolated from bovine mastitis: systematic review and meta-analysis. Prev Vet Med 2021; 188:105261. doi: 10.1016/j.prevetmed.2021.105261 [Crossref] [ Google Scholar]
- Algharib SA, Dawood A, Xie S. Nanoparticles for treatment of bovine Staphylococcus aureus mastitis. Drug Deliv 2020; 27(1):292-308. doi: 10.1080/10717544.2020.1724209 [Crossref] [ Google Scholar]
- Hashemi M, Kafi M, Safdarian M. The prevalence of clinical and subclinical mastitis in dairy cows in the central region of Fars province, south of Iran. Iran J Vet Res 2011; 12(3):236-41. doi: 10.22099/ijvr.2011.71 [Crossref] [ Google Scholar]
- Makovec JA, Ruegg PL. Results of milk samples submitted for microbiological examination in Wisconsin from 1994 to 2001. J Dairy Sci 2003; 86(11):3466-72. doi: 10.3168/jds.S0022-0302(03)73951-4 [Crossref] [ Google Scholar]
- Marlowe EM, Bankowski MJ. Conventional and molecular methods for the detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2011; 49(9 Suppl):S53-6. doi: 10.1128/jcm.00791-11 [Crossref] [ Google Scholar]
- Stuhlmeier R, Stuhlmeier KM. Fast, simultaneous, and sensitive detection of staphylococci. J Clin Pathol 2003; 56(10):782-5. doi: 10.1136/jcp.56.10.782 [Crossref] [ Google Scholar]
- Rocchetti TT, Martins KB, Martins PYF, de Oliveira RA, Mondelli AL, Fortaleza C. Detection of the mecA gene and identification of Staphylococcus directly from blood culture bottles by multiplex polymerase chain reaction. Braz J Infect Dis 2018; 22(2):99-105. doi: 10.1016/j.bjid.2018.02.006 [Crossref] [ Google Scholar]
- Koivula M, Pitkälä A, Pyörälä S, Mäntysaari EA. Distribution of bacteria and seasonal and regional effects in a new database for mastitis pathogens in Finland. Acta Agric Scand A Anim Sci 2007; 57(2):89-96. doi: 10.1080/09064700701488941 [Crossref] [ Google Scholar]
- Balemi A, Gumi B, Amenu K, Girma S, Gebru M, Tekle M. Prevalence of mastitis and antibiotic resistance of bacterial isolates from CMT positive milk samples obtained from dairy cows, camels, and goats in two pastoral districts in Southern Ethiopia. Animals (Basel) 2021; 11(6):1530. doi: 10.3390/ani11061530 [Crossref] [ Google Scholar]
- Zhang L, Li Y, Bao H, Wei R, Zhou Y, Zhang H. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microb Pathog 2016; 97:103-9. doi: 10.1016/j.micpath.2016.06.005 [Crossref] [ Google Scholar]
- Wielders CL, Fluit AC, Brisse S, Verhoef J, Schmitz FJ. mecA gene is widely disseminated in Staphylococcus aureus population. J Clin Microbiol 2002; 40(11):3970-5. doi: 10.1128/jcm.40.11.3970-3975.2002 [Crossref] [ Google Scholar]
- Pérez-Roth E, Claverie-Martín F, Villar J, Méndez-Alvarez S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol 2001; 39(11):4037-41. doi: 10.1128/jcm.39.11.4037-4041.2001 [Crossref] [ Google Scholar]
- Khazaie F, Ahmadi E. Bovine subclinical mastitis-associated methicillin-resistant Staphylococcus aureus, selective genotyping and antimicrobial susceptibility profile of the isolates in Kurdistan province of Iran. Iran J Microbiol 2021; 13(1):65-73. doi: 10.18502/ijm.v13i1.5494 [Crossref] [ Google Scholar]
- Ebrahimi A, Akhavan Taheri M. Characteristics of staphylococci isolated from clinical and subclinical mastitis cows in Shahrekord, Iran. Iran J Vet Res 2009; 10(3):273-7. doi: 10.22099/ijvr.2009.1708 [Crossref] [ Google Scholar]
- Ahangari Z, Ghorbanpoor M, Seifiabad Shapouri MR, Gharibi D, Ghazvini K. Methicillin resistance and selective genetic determinants of Staphylococcus aureus isolates with bovine mastitis milk origin. Iran J Microbiol 2017; 9(3):152-9. [ Google Scholar]
- Aarestrup FM, Jensen NE. Development of penicillin resistance among Staphylococcus aureus isolated from bovine mastitis in Denmark and other countries. Microb Drug Resist 1998; 4(3):247-56. doi: 10.1089/mdr.1998.4.247 [Crossref] [ Google Scholar]
- Matthews KR, Oliver SP, Jayarao BM. Susceptibility of staphylococci and streptococci isolated from bovine milk to antibiotics. Agri-Practice 1992; 13:18-24. [ Google Scholar]
- Martineau F, Picard FJ, Lansac N, Ménard C, Roy PH, Ouellette M. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 2000; 44(2):231-8. doi: 10.1128/aac.44.2.231-238.2000 [Crossref] [ Google Scholar]
- Sakoulas G, Gold HS, Venkataraman L, DeGirolami PC, Eliopoulos GM, Qian Q. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol 2001; 39(11):3946-51. doi: 10.1128/jcm.39.11.3946-3951.2001 [Crossref] [ Google Scholar]
- Varmazyar-najafi M, Pajohi-alamoti M, Mohammadzadeh A, Mahmoodi P. Detection of methicillin-resistance gene in Staphylococcus aureus isolated from traditional white cheese in Iran. Arch Hyg Sci 2016; 5(4):302-9. [ Google Scholar]