Avicenna Journal of Clinical Microbiology and Infection. 9(1):1-7.
doi: 10.34172/ajcmi.2022.01
Original Article
The Diagnostic Capacity of Three Phenotypic Techniques of Extended-Spectrum β-Lactamase Detection
Abera Abdeta 1, *  , Adane Bitew 2, Surafel Fentaw 1, Estifanos Tsige 1, Dawit Assefa 1, Eyasu Tigabu 3, Tadesse Lejisa 4, Yordanos Kefyalew 5, Ebisa Fekede 1
, Adane Bitew 2, Surafel Fentaw 1, Estifanos Tsige 1, Dawit Assefa 1, Eyasu Tigabu 3, Tadesse Lejisa 4, Yordanos Kefyalew 5, Ebisa Fekede 1
Author information:
1National Clinical Bacteriology and Mycology Reference Laboratory, Ethiopian Public Health Institute, Addis Ababa, Ethiopia
2Department of Medical Laboratory Sciences, College of Health Sciences, Addis Ababa University, Addis Ababa, Ethiopia
3Global One Health Initiative, The Ohio State University, East African Regional Office, Addis Ababa, Ethiopia
4National Clinical Chemistry Reference Laboratory, Ethiopian Public Health Institute, Addis Ababa, Ethiopia
5Department of Applied Biology, School of Applied Natural Science, Adama Science and Technology University, Adama, Ethiopia
*
Corresponding author: Abera Abdeta (MSc), P.O. Box: 1242 or 5654, Tel : +251911566420, Email:
aberaabdeta4@gmail.com
Abstract
Background: Early detection of extended-spectrum β-lactamases (ESBLs) producing bacteria is critical for infection prevention and control. Numerous phenotypic approaches and automated systems have been developed for detecting ESBL bacteria. However, there is a scarcity of data in Ethiopia regarding the most reliable, simple, and cost-effective methods for detecting ESBL-producing bacteria. This study, therefore, aimed to evaluate the diagnostic performance of three phenotypic approaches for detecting ESBL-producing bacteria.
Methods: In this study, 117 isolates of Klebsiella pneumoniae, Escherichia coli, Klebsiella oxytoca, and Proteus mirabilis were examined. Cefotaxime (30 µg) and ceftazidime (30 µg) were used for screening ESBL enzymes. A screening breakpoints of≤27 mm and≤22 mm were used for cefotaxime (30 µg) and ceftazidime (30 µg), respectively, as per the Clinical and Laboratory Standards Institute (CLSI) guidelines. All 117 strains were further confirmed by the Vitek 2 compact, double disk synergy, ESBL Epsilometer test, and combined disk method. The combined disk method was adopted as the reference method.
Results: Out of 117 isolates, 90 (86%) had zone diameters of≤27 mm and≤22 mm for cefotaxime (30 µg) and ceftazidime (30 µg), respectively. The reference method detected 76 (65%) ESBL isolates out of 117 ones. From among the three techniques (i.e., double disk synergy, Vitek 2 compact, and ESBL Epsilometer test), the double disk synergy method demonstrated overall sensitivity and specificity of 97.4% and 97.6%, respectively. Vitek-2, cefotaxime, and ceftazidime Epsilometer test indicated indeterminate results of 6.8%, 6.8%, and 5.1% respectively.
Conclusion: Double disk synergy was found to have the highest sensitivity and specificity for detecting ESBL isolates with no indeterminate results.
Keywords: Double disk synergy, Combined disk, Gradient strip, Vitek 2 compact
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Abdeta A, Bitew A, Fentaw S, Tsige E, Assefa D, Tigabu E, et al. The diagnostic capacity of three phenotypic techniques of extended-spectrum β-lactamase detection. Avicenna J Clin Microbiol Infect. 2022; 9(1):1-7. doi:10.34172/ajcmi.2022.01
Background
Extended-spectrum β-lactamases (ESBLs) are enzymes that are produced by bacteria causing resistance to a wide range of antibiotics including penicillins, cephalosporins, and aztreonam (1).Bacteria that produce ESBLs may also be resistant to unrelated and crucial antibiotics such as aminoglycosides, trimethoprim-sulfamethoxazole, and ciprofloxacin, making it difficult to treat critical patients (2).Oftentimes the infections caused by ESBL-producing Enterobacterales are associated with poor treatment outcomes (1,3).ESBLs hydrolyze the extended-spectrum cephalosporins and are becoming more common among Enterobacterales (4). The new breakpoints developed by European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Clinical and Laboratory Standards Institute (CLSI) for extended-spectrum cephalosporins have reduced the possibility of classifying an ESBL-producing Enterobacterales as susceptible to extended-spectrum cephalosporins (5,6). Therefore, identifying ESBL producing Enterobacterales is not compulsory for clinical outcome forecasting. However, for the purposes of epidemiological and infection prevention, it is very important to reduce its dissemination as well as monitor the distribution progress and the effectiveness of prevention strategies (5,6). Numerous phenotypic approaches have been developed for detecting ESBL-producing bacteria (7-13). The rising incidences of ESBLs-producing bacterial strains necessitate the development of low-cost, accurate, and simple testing techniques for detecting these enzymes in Enterobacterales isolates for epidemiological purposes. In Ethiopia, there is a scarcity of data regarding the most reliable, uncomplicated, and cost-effective techniques for the laboratory detection of extended-spectrum and lactamase-producing bacteria. As a result, the current study aimed to evaluate the diagnostic accuracy of three phenotypic approaches for detecting ESBL-producing bacteria.
Material and Methods
Study Design, Area, and Period
This cross-sectional study was conducted from January 5 to June 30, 2020, at the National Clinical Bacteriology and Mycology Reference Laboratory of Ethiopian Public Health Institute, in which Klebsiella pneumoniae, Escherichia coli, Klebsiella oxytoca, and Proteus mirabilis isolates recovered from clinical specimens were examined.
Sample Size Determination
The CLSI EP09-A3 recommended using at least 40 and preferably 100 samples to compare two laboratory methods. Therefore, a total of 117 above-mentioned isolates were collected in order to compare laboratory methods of ESBL detection (14).
Bacterial Culture and Identification
During the study period, 265 clinical samples were collected. In the event of sample transit delays, appropriate transport media were used. The isolates used in this investigation were recovered from blood, bodily fluids, wounds, sputum, and urine specimens. All specimens were inoculated onto appropriate culture media and cultured at proper temperatures and incubation time according to the per laboratory standard operating protocols (15). The isolates were identified by performing traditional biochemical tests: triple sugar iron agar (Liofilchem, Italy), oxidase strips (Liofilchem, Italy), Simon’s citrate agar (Liofilchem, Italy), and lysine iron agar (Liofilchem, Italy). Indole production and motility using sulfide-indole-motility medium (Liofilchem, Italy). Urease production using a urea agar base supplemented with 40% urea solution (Oxoid Ltd., England) (15). Also, 117 isolates of E. coli, K. pneumoniae, P. mirabilis, K. oxytoca isolates were isolated during the study period (Figure 1).
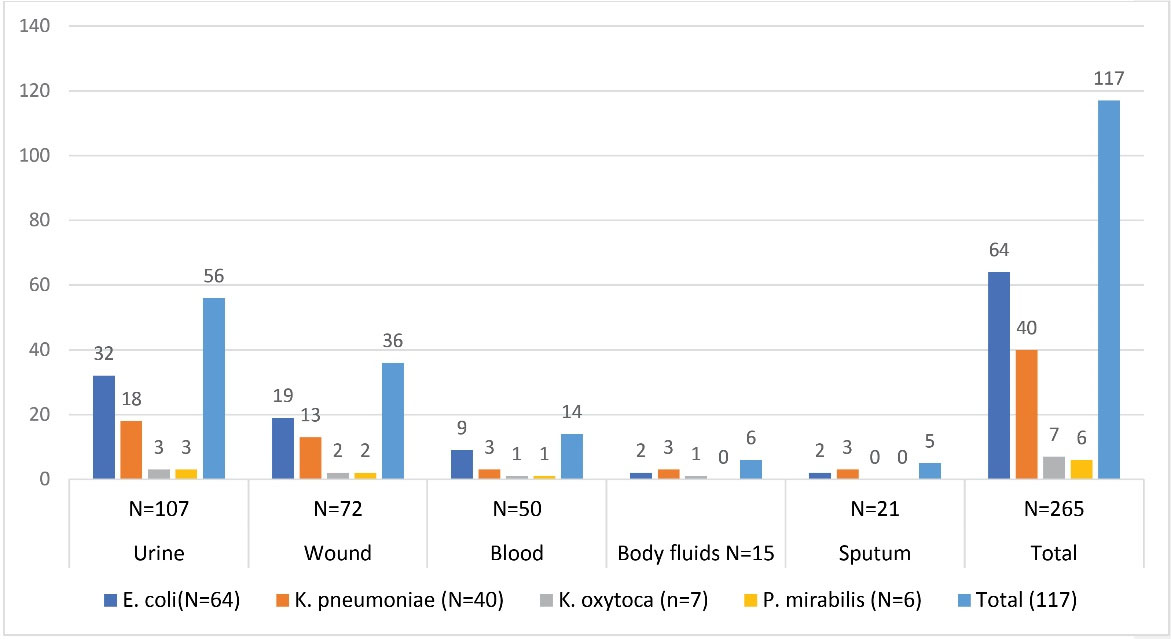
Figure 1.
Distribution of Different Isolates Among Clinical Specimens.
.
Distribution of Different Isolates Among Clinical Specimens.
Detection of Extended-Spectrum β-lactamases Producing Isolates
Screening of ESBL of Isolates
The disk diffusion method was adopted by using both cefotaxime (30 µg) (Oxoid Ltd., England) and ceftazidime (30 µg) (Oxoid Ltd., England) as indicator cephalosporins. A screening breakpoints of ≤ 27 mm and ≤ 22 mm were used for doing cefotaxime (30 µg) and ceftazidime (30 µg), respectively, on Mueller–Hinton agar (Oxoid Ltd, England) according to CLSI M100 guidelines (5). All 117 isolates were tested by confirmatory tests.
ESBL Phenotypic Confirmation Methods
Combined Disk Method (Reference Method)
The combined disk method was employed as the reference method. Ceftazidime 30 µg (Oxoid Ltd., England), Ceftazidime clavulanate (30/10 µg) (Oxoid Ltd., England), Cefotaxime 30 µg (Oxoid Ltd., England), and cefotaxime-clavulanate (30/10 µg) (Oxoid Ltd., England) were used on Mueller–Hinton agar in accordance with the CLSI guidelines 2020 (5) (Table 1). The plates were incubated at 37oC for 16–18 hours, and ESBL producers were defined as a 5-mm increase in zone diameter for indicator cephalosporins containing clavulanate vs indicator cephalosporins alone (5) (Figures 2 and 3). K. pneumoniae ATCC® 700 603 and E. coli ATCC® 25 922 control strains were used for checking the quality of all antibiotic disks.
Table 1.
Distribution of ESBL Producing Enterobacterales by Reference Method
|
Methods
|
|
E. coli
(n=64)
|
K. pneumoniae
(n=40)
|
K.
oxytoca
(n=7)
|
P. mirabilis
(n=6)
|
Total (117)
|
Percent
|
| CD CTX/CTL |
ND |
0 |
0 |
0 |
0 |
0 |
0.00 |
| Positive |
39 |
30 |
3 |
3 |
75 |
64.10 |
| Negative |
25 |
10 |
4 |
3 |
42 |
35.90 |
| CD CAZ/CAL |
ND |
0 |
0 |
0 |
0 |
0 |
0.00 |
| Positive |
40 |
30 |
3 |
3 |
76 |
65.00 |
| Negative |
24 |
10 |
4 |
3 |
41 |
35.00 |
Abbreviations: CAZ, ceftazidime; CTX, cefotaxime; ND, Indeterminate; ESBL, extended-spectrum β-lactamase.
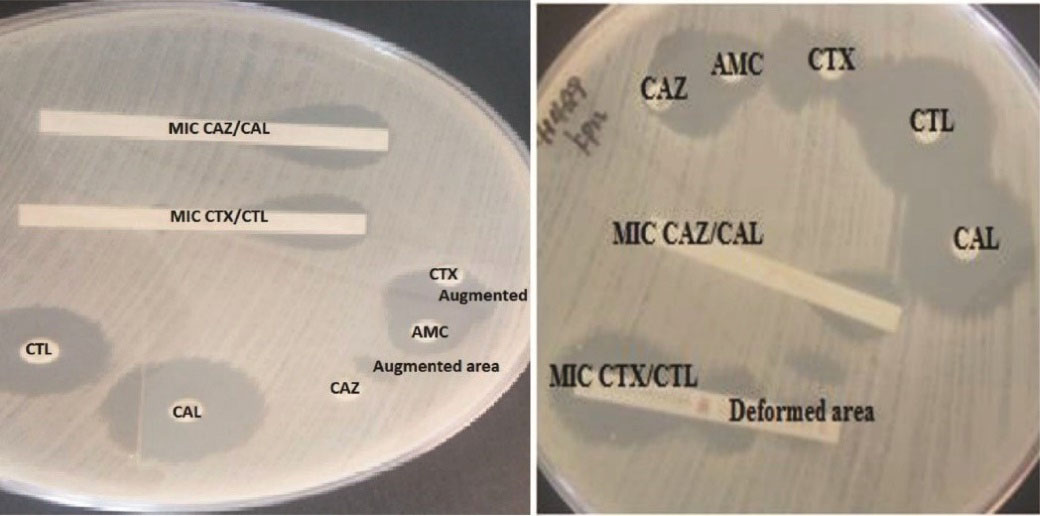
Figure 2.
Klebsiella Oxytoca(left) and Klebsiella Pneumoniae (Right) Positive for ESBL by DDS, ESBL E-test/MIC, and Combination Disk Methods on Muller Hinton Agar. Abbreviations: MIC, minimum inhibitory concentration; CAZ, ceftazidime; CTX, cefotaxime; CAL, ceftazidime/ clavulanic acid; CTL, cefotaxime/ clavulanic acid; AMC, amoxicillin/clavulanic acid; DDS, double-disk synergy.
.
Klebsiella Oxytoca(left) and Klebsiella Pneumoniae (Right) Positive for ESBL by DDS, ESBL E-test/MIC, and Combination Disk Methods on Muller Hinton Agar. Abbreviations: MIC, minimum inhibitory concentration; CAZ, ceftazidime; CTX, cefotaxime; CAL, ceftazidime/ clavulanic acid; CTL, cefotaxime/ clavulanic acid; AMC, amoxicillin/clavulanic acid; DDS, double-disk synergy.
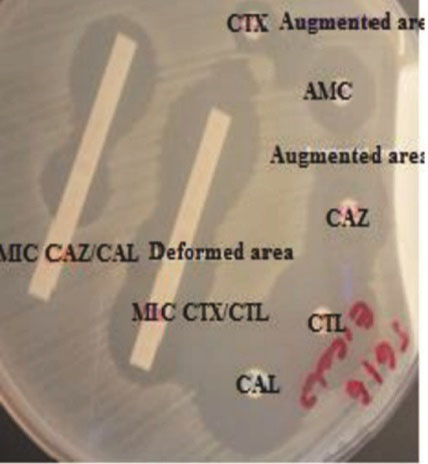
Figure 3.
ESBL Positive E. Coli by Double Disk Synergy, ESBL E-test/MIC, and Combination Disk Methods. Abbreviations: MIC, minimum inhibitory concentration; CAZ, ceftazidime; CTX, cefotaxime; CAL, ceftazidime/ clavulanic acid; CTL, cefotaxime/ clavulanic acid; AMC, amoxicillin/clavulanic acid; ESBL, Extended-spectrum β-lactamases.
.
ESBL Positive E. Coli by Double Disk Synergy, ESBL E-test/MIC, and Combination Disk Methods. Abbreviations: MIC, minimum inhibitory concentration; CAZ, ceftazidime; CTX, cefotaxime; CAL, ceftazidime/ clavulanic acid; CTL, cefotaxime/ clavulanic acid; AMC, amoxicillin/clavulanic acid; ESBL, Extended-spectrum β-lactamases.
Gradient Test Method/ Epsilometer Test
Strips containing cefotaxime/cefotaxime + clavulanic acid and ceftazidime/ceftazidime + clavulanic acid (Liofilchem, Italy) were used. Cefotaxime ≥0.5, cefotaxime/cefotaxime + clavulanic acid ratio ≥8 or CAZ ≥1, CAZ/CAL ratio ≥8 or distortion of the cefotaxime, and ceftazidime were considered ESBL positive. Cefotaxime < 0.5, or cefotaxime/cefotaxime+clavulanic acid ratio < 8, or ceftazidime < 1, or ceftazidime/ceftazidime + clavulanic acid ratio < 8 was considered ESBL negative. Cefotaxime > 16, cefotaxime + clavulanic acid > 1, ceftazidime > 32, and ceftazidime + clavulanic acid > 4 were considered inconclusive results (16) (Figures 2 and 3).
Double-disk Synergy Test (DDST)
Third-generation cephalosporins (cefotaxime and ceftazidime) were applied along with a third-generation cephalosporins disk containing clavulanic acid (amoxicillin-clavulanic acid) (Oxoid Ltd., England) (Oxoid Ltd., England). Enhanced inhibition zones around either of the disks in the direction of the clavulanic acid disk were considered ESBL positive. As for cephalosporin 30 µg disks, the gap between the disks was 20mm from center to center (6) (Figures 2 and 3).
VITEK 2 System ( bioMérieux )
VITEK 2 system antimicrobial susceptibility testing cards were used for doing (AST-GN86) (bioMérieux, Durham, North Carolina, USA). The results were analyzed using Vitek-2 compact software version 7.0, and The result was interpreted by the Vitek-2 compact software knowledge data base called the advanced expert system, which is used to interpret the antimicrobial susceptibility testing results.(17).
Quality Control
Klebsiella pneumoniae ATCC 700 603 and E. coli ATCC 25 922 Standard American type culture collectionstrainswere used as quality control strains.
Statistical Analysis
The detection power of each method was evaluated based on its sensitivity, specificity, and positive and negative predictive values. Cohen’s kappa value was used to measure the agreement between each phenotypic technique and reference method. Crosstabulation was used to calculate Cohen’s kappa value and P value. Cohen’s kappa values of 0.61-0.80 and 0.81-1.00 were used as good and very good agreement scores, respectively. P values ≤ 0.05 were considered statistically significant. The data were presented in tables and figures.
Results
In this study, 265 clinical specimens from five different clinical specimens were processed during the studied period. Out of the given number, 117 isolates of E. coli, K. pneumoniae, P. mirabilis, and K. oxytoca were recovered. E. coli was the most prevalent isolate, accounting for 64 (54.7%) of the isolates, followed by K. pneumoniae 40 (34.2%), K. oxytoca 7 (6%), and P. mirabilis 6 (5.1%). Out of 265 clinical specimens, most of the bacterial isolates were recovered from urine specimens (47.9%), then from wound (30.8%), blood (12%), body fluids (5%), and sputum (4.3%) (Figure 1). Out of 117 isolates, 90 (86%) had zone diameters of ≤ 27 mm and ≤ 22 mm for cefotaxime (30 µg) and ceftazidime (30 µg), respectively. Even though only 86 isolates showed reduced susceptibility to indicator cephalosporins (cefotaxime (30 µg) and ceftazidime (30 µg), all 117 isolates were tested by confirmatory tests. The reference method detected (n = 76/117, 65%) ESBL positive strains among 86 isolates with reduced susceptibility to cephalosporins (Table 1).
Vitek 2 Compact ESBL Results
Vitek 2 compact showed sensitivity (88.2%), specificity (82.9%), positive predictive value (90.5%), and negative predictive value (85%) with indeterminate results for 6.84% of the strains (Tables 2 and 3). Vitek 2 compact also showed a good measure of agreement with the reference method (kappa value = 0.709, P value = 0.001) (Table 3).
Table 2.
Distribution of ESBL Among Enterobacterales by Other Methods
|
Methods
|
|
E. coli
(n=64)
|
K. pneumoniae
(n=40)
|
K.
oxytoca
(n=7)
|
P. mirabilis
(n=6)
|
Total (117)
|
Percent
|
| DDS CAZ |
ND |
0 |
0 |
0 |
0 |
0 |
0 |
| Positive |
39 |
30 |
3 |
3 |
75 |
64.1 |
| Negeative |
25 |
10 |
4 |
3 |
42 |
35.9 |
| DDS CTX |
ND |
0 |
0 |
0 |
0 |
0 |
0 |
| Positive |
39 |
30 |
2 |
3 |
74 |
63.2 |
| Negeative |
25 |
10 |
5 |
3 |
43 |
36.8 |
| ESBL E CAZ |
ND |
1 |
3 |
1 |
1 |
6 |
5.1 |
| Positive |
37 |
28 |
3 |
3 |
71 |
60.7 |
| Negeative |
26 |
9 |
3 |
2 |
40 |
34.2 |
| ESBL E CTX |
ND |
2 |
3 |
2 |
1 |
8 |
6.8 |
| Positive |
37 |
27 |
3 |
3 |
70 |
59.8 |
| Negeative |
25 |
10 |
2 |
2 |
39 |
33.4 |
| ESBL VITEK 2 AST GN-86 |
ND |
3 |
2 |
2 |
1 |
8 |
6.8 |
| Positive |
37 |
27 |
3 |
3 |
70 |
59.8 |
| Negeative |
25 |
10 |
2 |
2 |
39 |
33.4 |
Abbreviations: DDS, double-disk synergy; CAZ, Ceftazidime; CTX, Cefotaxime; ND, Indeterminate; E; Epsilometer; ESBL, Extended-spectrum β-lactamase.
Table 3.
Statistical Analysis of Different Phenotypic Methods of ESBL Detection
|
Phenotypic Methods
|
%Sensitivity
|
%Specificity
|
%PPV
|
%NPV |
Kappa Value
|
P
Value
|
| DDS CAZ |
97.4 |
97.6 |
98.7 |
95.2 |
0.944 |
0.001 |
| DDS CTX |
96.1 |
97.6 |
98.6 |
93.8 |
0.926 |
0.001 |
| Vitek 2 |
88.2 |
82.9 |
90.5 |
85 |
0.709 |
0.001 |
| ECAZ |
93.4 |
95.1 |
100 |
97.5 |
0.877 |
0.001 |
| ECTX |
90.8 |
92.7 |
98.6 |
97.4 |
0.827 |
0.001 |
Abbreviations: DDS, double-disk synergy; CAZ, ceftazidime; CTX, cefotaxime; E; Epsilometer; PPV, positive predictive value; NPV, negative predictive value.
Double-Disk Synergy Test
DDST using ceftazidime and amoxicillin with clavulanic acid showedsensitivity (97.4%), specificity (97.6%), positive predictive value (98.7%), and negative predictive value (95.2%). DDST using cefotaxime and amoxicillin with clavulanic acid showed sensitivity (96.1%), specificity (97.6%), positive predictive value (98.6%), and negative predictive value (93.8%) (Table 3). The double-disk synergy showed a very good measure of agreement with the reference method (kappa value = 0.926-0.944, P value = 0.001) (Tables 2 and 3).
ESBL Detection Using E-test
The ESBL E-test ceftazidime/ceftazidime + clavulanic acid strips showed sensitivity, specificity, positive predictive value, and negative predictive values of 93.4%, 95.1%, 100%, and 97.5%, respectively, and with indeterminate results for 5.1% of the strains. The sensitivity, specificity, positive predictive value, and negative predictive value of the cefotaxime/cefotaxime+clavulanic acid strips were 90.8%, 92.7%, 98.6%, and 97.4%, respectively, and with indeterminate results for 6.84% of the strains (Tables 2 and 3). The ESBL E-tests showed a very good measure of agreement with the reference method (kappa value = 0.827-0.877, P value = 0.001) (Tables 2 and 3).
Discussion
The new extended-spectrum cephalosporin and aztreonam breakpoints reduced the possibility of determining ESBL producers susceptible to these antibiotics. However, timely detection of ESBL-producing Enterobacterales is also very important for early designing and implementing control strategies (1,2).
Vitek 2 Compact System
In our study, the Vitek 2 compact method demonstrated a sensitivity and specificity of 88.2% and 82.9%, respectively. This result was in line with the findings from Wiegand et al study in which the sensitivity and specificity were 86% and 78%, respectively (13), and from Thomson et al study in which the sensitivity and specificity were 89% and 85%, respectively (18). However, following researchers reported sensitivity results that were comparable to ours, but documented higher specificity results: Singh et al: sensitivity 91.8% and specificity 97.24 % (19); Robin et al: sensitivity 91.8% and specificity 100% (20); and Young et al: sensitivity 92% and specificity 100% (21). The following researchers, on the other hand, reported higher sensitivity and specificity for Vitek 2 compact: Spanu et al: sensitivity 98.1% and specificity 99.7% (10); Hackman et al: sensitivity 98.5% and specificity 98.9% (22); Sorlózano et al: sensitivity 100% and specificity 99.3% (23); and Valenza et al: sensitivity 100% and specificity 96% (24). Garrec et al reported sensitivity and specificity of 50% to 79% (25) for different Vitek 2 cards, which were much lower than those found in our study. This inconsistency may have been attributed to the differences in geography, selected bacterial isolate groups, and adopted Vitek 2 compact cards.
ESBL E-tests
According to our study results, the sensitivity and specificity for ESBL E-tests were 92% and 98%, respectively, when both MIC ceftazidime and cefotaxime were used. Our results in this regard were consistent with findings from the studies by following researchers: Linscott and Brown: sensitivity 97% and specificity 94% (26); Singh et al: sensitivity 88.52% and specificity 100% (18); Brown et al: sensitivity 88.9% and specificity 100% (27); Cormican et al: 100% for both sensitivity and specificity (7); Ho et al: sensitivity 96% and specificity 100% (28); and Sorlózano et al: sensitivity 100% and specificity 99.3%. Our results, however, were only partially consistent with findings from the studies by following researchers since they reported relatively lower sensitivity: Vercauteren et al: sensitivity 84% and specificity 100% (29); and Yang et al ESBL E-test: sensitivity 84.0% and specificity 100% (30). The inconsistency between the results may have been attributable to the utilized bacterial isolates, since we only used K. pneumoniae, K. oxytoca, P. mirabilis, and E. coli, unlike other researchers, the geographical difference. In the current study, ESBL E-test yielded indeterminate results of 5.1% to 6.8%, which were lower than the results in a study by Garrec et al (28) reporting indeterminate results of 11% to 49% for the ESBL E-test. This inconsistency may have been explained by the fact that they utilized other organisms in addition to K. pneumoniae, E. coli, K. oxytoca, and P. mirabilis as well as employed MIC cefepime strips, in addition to MIC ceftazidime and cefotaxime strips.
Double-Disk Synergy Methods
In this study, the overall sensitivity and specificity of the DDST were detected to be 97.37% and 97.56%, respectively. Our findings are partially consistent with the findings of other researchers who investigated the diagnostic capacity of double disk synergy methods using ESBL and non-ESBL producing bacteria (ranging from 79% to 97% and 94% to 100%, respectively), which were initially confirmed using PCR methods(11,23,24,31,32). However, our study findings contradicted the results from the studies by Yang et al reporting sensitivity of 96.0% and specificity of 69.2% for the double-disk synergy method (25),andby De Gheldre et al determining sensitivity of 89% and specificity of 92% (33). The observed inconsistency was likely due to the different Enterobacterales strains used (in this study, only K. pneumoniae, K. oxytoca, P. mirabilis, and E. coli were used, whereas K. aerogenes, P. vulgaris, Providencia stuartii, and others were applied in the studies by other researchers), regional differences, and so on.
Conclusion
In sum, Vitek-2 compact and ESBL E cefotaxime/cefotaxime with clavulanic acid yielded inconclusive findings of 6.8 percent. In addition, ESBL E ceftazidime/ceftazidime with clavulanic acid produced 5.1 percent indeterminate findings. However, the double-disk synergy method was shown to detect more ESBL producers with no indeterminate results. Therefore, it was concluded that the double-disk synergy method was more reliable, simpler, and less expensive method which also required no specialized equipment or considerable expertise.
Acknowledgments
The authors would like to appreciate the Ethiopian Public Health Institute for donating supplies for this study, as well as Dr. Adane Bitew and Dr. Eyasu Tigabu for their assistance.
Conflict of Interests
The authors declare that they have no conflict of interests.
Ethical Approval
The Ethical clearance of this study was obtained from the Ethical Review Committee of the Department of Medical Laboratory Sciences, College of Health Sciences, Addis Ababa University (DRERC/474/19/MLS). In addition, the Ethiopian Public Health Institute granted an official permission.
References
- Bush K, Jacoby GA, Medeiros AA. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother 1995; 39(6):1211-33. doi: 10.1128/aac.39.6.1211 [Crossref] [ Google Scholar]
- Jacoby GA, Medeiros AA. More extended-spectrum beta-lactamases. Antimicrob Agents Chemother 1991; 35(9):1697-704. doi: 10.1128/aac.35.9.1697 [Crossref] [ Google Scholar]
- Munoz-Price LS, Jacoby GA, Snydman DR. Extended-Spectrum Beta-Lactamases. UpToDate online; 2019.
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 2001; 14(4):933-51. doi: 10.1128/cmr.14.4.933-951.2001 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100. Wayne, PA: CLSI; 2021.
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). EUCAST Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance. Basel, Switzerland: EUCAST; 2017. http://www.eucast.org/clinical_breakpoints.
- Cormican MG, Marshall SA, Jones RN. Detection of extended-spectrum beta-lactamase (ESBL)-producing strains by the E-test ESBL screen. J Clin Microbiol 1996; 34(8):1880-4. doi: 10.1128/jcm.34.8.1880-1884.1996 [Crossref] [ Google Scholar]
- Donaldson H, McCalmont M, Livermore DM, Rooney PJ, Ong G, McHenry E. Evaluation of the VITEK 2 AST N-054 test card for the detection of extended-spectrum beta-lactamase production in Escherichia coli with CTX-M phenotypes. J Antimicrob Chemother 2008; 62(5):1015-7. doi: 10.1093/jac/dkn316 [Crossref] [ Google Scholar]
- Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis 1988; 10(4):867-78. doi: 10.1093/clinids/10.4.867 [Crossref] [ Google Scholar]
- Spanu T, Sanguinetti M, Tumbarello M, D’Inzeo T, Fiori B, Posteraro B. Evaluation of the new VITEK 2 extended-spectrum beta-lactamase (ESBL) test for rapid detection of ESBL production in Enterobacteriaceae isolates. J Clin Microbiol 2006; 44(9):3257-62. doi: 10.1128/jcm.00433-06 [Crossref] [ Google Scholar]
- Thomson KS, Sanders CC. Detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae: comparison of the double-disk and three-dimensional tests. Antimicrob Agents Chemother 1992; 36(9):1877-82. doi: 10.1128/aac.36.9.1877 [Crossref] [ Google Scholar]
- Tzelepi E, Giakkoupi P, Sofianou D, Loukova V, Kemeroglou A, Tsakris A. Detection of extended-spectrum beta-lactamases in clinical isolates of Enterobacter cloacae and Enterobacter aerogenes. J Clin Microbiol 2000; 38(2):542-6. doi: 10.1128/jcm.38.2.542-546.2000 [Crossref] [ Google Scholar]
- Wiegand I, Geiss HK, Mack D, Stürenburg E, Seifert H. Detection of extended-spectrum beta-lactamases among Enterobacteriaceae by use of semiautomated microbiology systems and manual detection procedures. J Clin Microbiol 2007; 45(4):1167-74. doi: 10.1128/jcm.01988-06 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Measurement Procedure Comparison and Bias Estimation Using Patient Samples; Approved Guideline. 3rd ed. CLSI document EP09-A3. Wayne, PA: CLSI; 2013.
- Leber AL. Clinical Microbiology Procedures Handbook. 4th ed. John Wiley & Sons; 2016.
- Cefotaxime C, Ctl CTX, Cal CAZ, Cal CAZ, Cal CAZ, Cal CAZ. MIC Test Strip Technical Sheet ESBL 2014;2–5.
- Tips & Tricks for the Advanced Expert System. https://www.biomerieux-microbio.com/tips-tricks-for-the-advanced-expert-system-aes/.
- Singh RM, Singh HL. Comparative evaluation of six phenotypic methods for detecting extended-spectrum beta-lactamase-producing Enterobacteriaceae. J Infect Dev Ctries 2014; 8(4):408-15. doi: 10.3855/jidc.4052 [Crossref] [ Google Scholar]
- Hackman HK, Osei-Adjei G, Gordon A, Laryea E, Quaye S, Anison L. The reliability of using Vitek 2 compact system to detect extended-spectrum beta-lactamaseproducing Isolates in Escherichia coli and Klebsiella pneumoniae in Accra, Ghana. Adv Life Sci Technol 2013; 13:84-90. [ Google Scholar]
- Linscott AJ, Brown WJ. Evaluation of four commercially available extended-spectrum beta-lactamase phenotypic confirmation tests. J Clin Microbiol 2005; 43(3):1081-5. doi: 10.1128/jcm.43.3.1081-1085.2005 [Crossref] [ Google Scholar]
- Brown DF, Andrews J, King A, MacGowan AP. Detection of extended-spectrum beta-lactamases with E-test and double-disc potentiation methods. J Antimicrob Chemother 2000; 46(2):327-8. doi: 10.1093/jac/46.2.327 [Crossref] [ Google Scholar]
- Ho PL, Chow KH, Yuen KY, Ng WS, Chau PY. Comparison of a novel, inhibitor-potentiated disc-diffusion test with other methods for the detection of extended-spectrum beta-lactamases in Escherichia coli and Klebsiella pneumoniae. J Antimicrob Chemother 1998; 42(1):49-54. doi: 10.1093/jac/42.1.49 [Crossref] [ Google Scholar]
- Vercauteren E, Descheemaeker P, Ieven M, Sanders CC, Goossens H. Comparison of screening methods for detection of extended-spectrum beta-lactamases and their prevalence among blood isolates of Escherichia coli and Klebsiella spp in a Belgian teaching hospital. J Clin Microbiol 1997; 35(9):2191-7. doi: 10.1128/jcm.35.9.2191-2197.1997 [Crossref] [ Google Scholar]
- Yang JL, Wang JT, Lauderdale TL, Chang SC. Prevalence of extended-spectrum beta-lactamases in Enterobacter cloacae in Taiwan and comparison of 3 phenotypic confirmatory methods for detecting extended-spectrum beta-lactamase production. J Microbiol Immunol Infect 2009; 42(4):310-6. [ Google Scholar]
- Bedenic B, Randegger C, Boras A, Haechler H. Comparison of five different methods for detection of SHV extended-spectrum beta-lactamases. J Chemother 2001; 13(1):24-33. doi: 10.1179/joc.2001.13.1.24 [Crossref] [ Google Scholar]
- MacKenzie FM, Miller CA, Gould IM. Comparison of screening methods for TEM- and SHV-derived extended-spectrum beta-lactamase detection. Clin Microbiol Infect 2002; 8(11):715-24. doi: 10.1046/j.1469-0691.2002.00473.x [Crossref] [ Google Scholar]
- Garrec H, Drieux-Rouzet L, Golmard JL, Jarlier V, Robert J. Comparison of nine phenotypic methods for detection of extended-spectrum beta-lactamase production by Enterobacteriaceae. J Clin Microbiol 2011; 49(3):1048-57. doi: 10.1128/jcm.02130-10 [Crossref] [ Google Scholar]
- Thomson KS, Cornish NE, Hong SG, Hemrick K, Herdt C, Moland ES. Comparison of Phoenix and VITEK 2 extended-spectrum-beta-lactamase detection tests for analysis of Escherichia coli and Klebsiella isolates with well-characterized beta-lactamases. J Clin Microbiol 2007; 45(8):2380-4. doi: 10.1128/jcm.00776-07 [Crossref] [ Google Scholar]
- Robin F, Delmas J, Schweitzer C, Bonnet R. Evaluation of the Vitek-2 extended-spectrum beta-lactamase test against non-duplicate strains of Enterobacteriaceae producing a broad diversity of well-characterised beta-lactamases. Clin Microbiol Infect 2008; 14(2):148-54. doi: 10.1111/j.1469-0691.2007.01893.x [Crossref] [ Google Scholar]
- Young AL, Nicol MP, Moodley C, Bamford CM. The accuracy of extended-spectrum beta-lactamase detection in Escherichia coli and Klebsiella pneumoniae in South African laboratories using the Vitek 2 Gram-negative susceptibility card AST-N255. S Afr J Infect Dis 2019; 34(1):114. doi: 10.4102/sajid.v34i1.114 [Crossref] [ Google Scholar]
- Sorlózano A, Gutiérrez J, Piédrola G, Soto MJ. Acceptable performance of VITEK 2 system to detect extended-spectrum beta-lactamases in clinical isolates of Escherichia coli: a comparative study of phenotypic commercial methods and NCCLS guidelines. Diagn Microbiol Infect Dis 2005; 51(3):191-3. doi: 10.1016/j.diagmicrobio.2004.11.001 [Crossref] [ Google Scholar]
- Valenza G, Müller S, Schmitt C, Turnwald D, Lam TT, Frosch M. Evaluation of the VITEK 2 AST-N111 card for detection of extended-spectrum beta-lactamases (ESBLs) in Escherichia coli, Klebsiella pneumoniae, and Klebsiella oxytoca compared to ESBL E-tests and combination disk methods. Eur J Clin Microbiol Infect Dis 2011; 30(7):869-72. doi: 10.1007/s10096-011-1169-2 [Crossref] [ Google Scholar]
- De Gheldre Y, Avesani V, Berhin C, Delmée M, Glupczynski Y. Evaluation of Oxoid combination discs for detection of extended-spectrum beta-lactamases. J Antimicrob Chemother 2003; 52(4):591-7. doi: 10.1093/jac/dkg415 [Crossref] [ Google Scholar]