Avicenna Journal of Clinical Microbiology and Infection. 9(1):8-15.
doi: 10.34172/ajcmi.2022.02
Original Article
Isolation of Actinobacteria Strains From Environmental Samples and Assessment of Their Bioactivity
Mehdi Mohammadi 1  , Moj Khaleghi 1, *
, Moj Khaleghi 1, *  , Shahriyar Shakeri 2, Majid Askari Hesni 1, Mohammad Rasoul Samandari-Bahraseman 3, Ava Dalvand 1
, Shahriyar Shakeri 2, Majid Askari Hesni 1, Mohammad Rasoul Samandari-Bahraseman 3, Ava Dalvand 1 
Author information:
1Department of Biology, Faculty of Sciences, Shahid Bahonar University of Kerman, Kerman, Iran
2Department of Biotechnology, Institute of Science and High Technology and Environmental Sciences, Graduate University of Advanced Technology, Kerman, Iran
3Department of Production Engineering and Plant Genetics, Faculty of Agriculture, Lorestan University, Khorramabad, Iran
*
Corresponding author: Moj Khaleghi, Tel: +983431322046, Fax: +983433257165, Email:
m.khaleghi@uk.ac.ir
Abstract
Background:
Actinobacteria are widespread and live in a variety of habitats. Today, these bacteria are very important due to the production of various secondary metabolites with different biological activities. The present study aimed to isolate strains of Actinobacteria from different habitats (the Persian Gulf, Gandom Beryan area in the Lut Desert, and some plant roots). The anticancer and antimicrobial activities of secondary metabolites of these isolates were also investigated.
Methods: Samples were taken from water of the Persian Gulf, soil of Gandom Beryan area in the Lut Desert, and plant roots. For isolation of Actinobacteria, samples were cultured in ISP2, ISP4, AIA, Gauze, M1, ISP3, and GYP media. Bacterial strains were identified based on the colony and bacterial morphology and confirmed using the specific primers for Actinobacteria. The anticancer and antimicrobial activities of crude metabolite extracts and supernatant of the isolates were evaluated on MCF-7 and Staphylococcus aureus PTCC 112 and Pseudomonas aeruginosa PTCC 1214 strains.
Results: The results showed that the supernatants of 7 isolates (ga31, ez, sa, mar2, rz, ga33, and ga5) and the metabolite extracts of 4 strains (ga31, ga5, rz, and ez) had anticancer activity. Overall, ga31 was the best strain with anticancer activity of more than 75%. When evaluating the antimicrobial activity of bacterial secondary metabolites, we found that only two strains (ga31 and ga5) had antimicrobial activity against S. aureus PTCC 1112.
Conclusions: In general, strain ga31, which has high anticancer and antimicrobial activities, could be a good candidate for new trials in the pharmaceutical industry.
Keywords: Actinobacteria, Anticancer, Antimicrobial, Secondary metabolite, Persian Gulf
Copyright and License Information
© 2022 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Please cite this article as follows: Mohammadi M, Khaleghi M, Shakeri S, Askari Hesni M, Samandari-Bahraseman MR, Dalvand A. Isolationof actinobacteria strains from environmental samples and assessment of their bioactivity. Avicenna J Clin Microbiol Infect. 2022; 9(1):8-15.doi:10.34172/ajcmi.2022.02
Background
Actinobacteria are gram-positive and aerobic bacteria with a high C + G content in their genome. They are important producers of natural bioactive metabolites, such as antibiotics, anticancer compounds, antifungal compounds, enzymes, immunosuppressants, and metabolites with broad industrial applications (1,2). In recent years, Actinobacteria have received greater attention for the isolation and identification of new antimicrobial and anticancer compounds due to the increased resistance of microorganisms to drugs against antibiotics placed on the market, as well as many side effects of existing chemotherapeutic drugs (3-5).
The Actinomycetaceae family, as the main source of antimicrobial agents, is one of the most important members of the Actinobacteria class (6). They are adapted to a variety of aquatic, terrestrial, and aerial environments. Their density and types in these habitats are affected by environmental factors such as temperature, nutrients, and pH. However, the soil is known as the main habitat of these bacteria. On the other hand, the abundance of Actinobacteria in the soil has made them have a high yield in interaction with plants (7,8). Besides, marine environments are also known to be the most important habitats for these bacteria. Therefore, in recent years, they have received a lot of attention due to the production of biological metabolites (9). Based on past studies, the researchers have found that different environments can be habitats for different species of Actinobacteria that have not yet been identified but can be rich sources of bioactive components (10). In recent years, the discovery of new bioactive compounds from various soil bacteria has decreased, and this may be due to the effect of saturation and the non-isolation of new strains. Therefore, isolating and identifying different bacterial strains (including Actinobacteria) from different habitats may increase the chances of new metabolites being discovered. For this reason, scientists recommend that isolating these bacteria from unusual environments with specific environmental conditions, such as marine and desert ecosystems, could be a way to discover new strains and novel valuable chemical compounds (3,11).
Consequently, in this study, we attempted to isolate various strains of Actinobacteria from different ecosystems (the Persian Gulf, Gandom Beryan area in the Lut Desert, and plant roots) and investigate the biological activities of their metabolites, including anticancer and antimicrobial activities.
Materials and Methods
Sampling and Isolation of Actinobacteria
Actinobacteria strains were isolated from soil, water, and plant roots. Samples were collected from three sources (water of Persian Gulf, the soil of Gandom Beryan area in the Lut Desert, and plant roots) in sterile bottles and jars and were taken to the laboratory immediately. To isolate different strains of Actinobacteria, first, the samples were placed in the water bath at 55°C for 60 minutes. Then, serial dilutions (10-1 -10-4) were prepared and 100 µL of each dilution was cultured on agar media (ISP2, ISP4, AIA, Gauze, M1, ISP3, and GYP) (3). Nalidixic acid (10 μg/mL) and nystatin (50 μg/mL) were added to each culture medium to avoid bacterial and fungal contamination (12). After inoculation, the plates were incubated for 21 days at 28°C. Then, the powdery colonies were chosen for future investigation. The isolated strains were stored in a culture medium containing glycerol (20%, v/v) at -20°C for subsequent experiments.
Identification of Isolated Bacteria
Initial identification of Actinobacteria was performed by morphological studies. Colony and bacterial morphology were investigated by light microscopy after 21 days at 28°C at magnifications of 100× (Zeiss Stemi 508) and 1000× (Olympus Microscope CX43), respectively. Then, the identification of the isolated strains was done by molecular tests. Finally, the isolates were examined through biochemical tests (including Voges-Proskauer and Methyl red, citrate, urease, ornithine, lysine, indole, nitrate, arginine, phenylalanine-deamination, triple sugar iron agar, starch hydrolysis, and carbon source utilization) (13,14).
For final approval, the molecular study was done using the class-specific primer (Sm6F: (5́-GGTGGCGAAGGCGGA-3́), Sm5R (5́-GAACTGAGACCGGCTTTTTGA-3́), and S-C-Act-235-a-S-20: F (5́- CGCGGCCTATCAGCTTGTTG-3́), S-C-Act-878-a-A-19: R (5́ CCGTACTCCCCAGGCGGGG-3́) (15). Genomic DNA was extracted using bacterial DNA isolation kit (DENA Zist Asia, Iran) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) program consisted of an initial denaturation at 94°C for 3 minutes, 35 cycles of denaturation at 94°C for 1 minute, annealing at 55°C for 1 minute, extension at 72°C for 1 minute, and final extension at 72°C for 10 minutes (16). The PCR product was purified using a PCR purification kit (Thermo Scientific) and was checked by 1% agarose gel. The 600-640 bp bands indicated that the isolated strains belonged to the Actinobacteria phylum.
Finally, the 16s rRNA sequence of the selected primer was determined by universal primer (PA: F (5́ AGAGTTTGATCCTGGCTCAG-3), PH: R (5́-AAGGAGGTGATCCAGCCGCA-3́) (17). The PCR program was carried out according to the above-mentioned method. The PCR product was purified by AccuPrep® PCR Purification gel (Bioneer, South Korea) according to the manufacturer’s instructions. Then, DNA sequencing was performed by the South Korean company Bioneer. The sequence data of 16S rRNA were then utilized for BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) analysis and the maximum likelihood phylogenetic tree was constructed using the Molecular Evolutionary Genetics Analysis software version 4.0 (18).
Preparation of Crude Metabolites Extracts for bioactivity Assessment
For the assessment of the bioactivity of the isolated Actinobacteria, each strain was inoculated in 200 mL of proprietary medium, incubated at 37°C with 140 rpm for 8 days, and then centrifuged at 10000 rpm for 10 minutes at 30°C (5). The supernatant was collected and filtered with a 0.22 µm filter. Finally, for the extraction of bacterial metabolites, the supernatant was mixed with an equal amount (1:1 V/V) of ethyl acetate (Merck, Germany) and shaken vigorously for 10 minutes. Then, the organic phase was extracted. The extracts were then dried by rotary evaporation and maintained at -20°C (19,20).
Assessment of Anticancer Activity of Crude Metabolite Extract and Supernatant
The anticancer activity of crude extract and supernatant of isolated strains was investigated on the MCF-7 cell line (from the Iranian Biological Resource Center) using the MTT method. MTT test was done based on the method used by Azman et al in 2017 (21). At first, the cancer cells at a density of 5×102 in 96 well microtiter plates were incubated in Dulbecco’s modification of Eagle’s medium (DMEM) supplemented with glucose, 10% fetal bovine serum, and antibiotics (20 μg/mL of Penicillin and 100 μg/mL of Streptomycin) at 37°C in 5% carbon dioxide for 24 hours (22). Then, 20 µL of each bacterial sample (crude metabolite extract and supernatant) was added to each well separately (final concentration of 30 µg/mL) and incubated for 24 hours. After the incubation period, 20 μL of MTT solution was added to each of the wells and the plates were incubated for 4 hours in the darkness. After this time, the cell culture media were removed, and 100 μL of DMSO was added to each of the wells. The optical absorption was measured by Elisa reader (Lab biotech, USA) at the wavelength range of 490-690 nm and the following equation was used to calculate the percentage of cell viability:
Cell viability (%) = (Average Optical absorption for treated cells/ Average optical absorption of control cells) × 100
The control sample was treated with DMSO solvent (0.5%).
Assessment of the Antimicrobial Activity of Crude Metabolite Extract and Supernatant
The antimicrobial activity of crude bacterial extract and supernatant of the isolated strains was investigated against Staphylococcus aureus PTCC 1112 and Pseudomonas aeruginosa PTCC 1214 using the disk diffusion method (23). The bacterial pathogens were cultured in the Mueller Hinton Broth medium. Then, the 0.5 McFarland turbidity standard was prepared from each isolate and spread on Mueller Hinton agar medium. The sterile blank disc (6 mm diameter) (Padtan Teb, Iran) containing 20 μL of the supernatant and crude bacterial extract (1 mg/mL) was placed on the culture medium. After incubation at 37°C for 24 hours, the inhibition zone (mm) was measured. Gentamycin disc (10 μg) and the blank disc with DMSO (5%) were used as the positive and negative control, respectively.
Determination of Minimum Inhibitory and Bactericidal Concentrations
Determination of the minimum inhibitory and bactericidal concentrations of crude extract and supernatant of isolated strains were conducted based on the method used by Weseler et al (24) with some modifications. The serial dilutions (1.95 µg/mL to 1 mg/mL) were prepared from each of the crude extracts and supernatant of bacteria, separately. Afterwards, 100 μL of each sample was added to the wells. Then, 20 μL of pathogenic bacteria (0.5 McFarland turbidity) was also added and incubated at 37°C for 18-24 hours. Afterwards, the turbidity of each well was assessed. Finally, to determine the minimum inhibitory and bactericidal concentrations of bacterial products, 10 μL of the first clear well was inoculated into the Mueller Hinton agar. After the incubation at 37°C for 24 hours, the growth or non-growth of pathogenic bacteria was investigated. MIC is defined as the lowest concentration of an antimicrobial that can inhibit the visible growth of a microorganism after incubation, while MBC is the lowest concentration of antibacterial agent required to kill a bacterium.
The culture medium without bacterial products was used as the negative control.
Statistical Analysis
All experiments were performed with three repetitions. Data analysis was performed using one-way analysis of variance (ANOVA) in SPSS version 21.0 with a significance level of P ≤ 0.05.
Results
Isolation of Actinobacteria
Based on morphological and microscopic characteristics, 112 Actinobacteria strains were identified in all studied sources (Figure 1), which were grown on six culture media (Table 1). The majority of isolated strains belonged to Gauze and ISP4. Molecular identification using the class-specific primers showed that the isolated strains belonged to Actinobacteria phylum, which was confirmed by the appearance of 600-640 bp bands on the agarose gel (Figure 2).

Figure 1.
The Actinobacteria Strains Isolated from Seawater, Soil, and Plant Root samples. (A, B) Primary cultivation from different samples of seawater, plant roots, and soil, (C-L) Pure cultivation from isolated strains, (M-P) Morphology, and the gram reaction of isolated bacteria (magnification 1000×).
.
The Actinobacteria Strains Isolated from Seawater, Soil, and Plant Root samples. (A, B) Primary cultivation from different samples of seawater, plant roots, and soil, (C-L) Pure cultivation from isolated strains, (M-P) Morphology, and the gram reaction of isolated bacteria (magnification 1000×).
Table 1.
Actinobacteria Strains Isolated from Different Sources
|
Sample
|
Culture Media
|
|
SCA
|
ISP2
|
ISP4
|
Gauze
|
AIA
|
GYP
|
| Sea |
2 |
3 |
1 |
1 |
1 |
2 |
| Soil |
10 |
12 |
21 |
27 |
17 |
12 |
| Plant root |
- |
- |
1 |
1 |
1 |
- |
| Total |
12 |
15 |
23 |
29 |
19 |
14 |
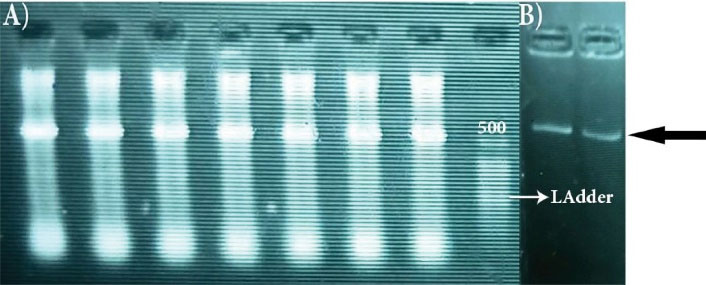
Figure 2.
(A) Observation of 600-640 bp bands marked with the black arrow. (B) DNA ladder 25 bp.
.
(A) Observation of 600-640 bp bands marked with the black arrow. (B) DNA ladder 25 bp.
Biochemical tests exhibited 22 isolated strains with different biochemical properties (data not shown). In Table 2, some morphological properties of these isolates were exhibited.
Table 2.
Actinobacteria Strains Isolated FROM Seawater, Soil, and Plant Roots
|
Culture Media
|
Pigment
|
Substrate Mycelium
|
Aerial Mycelium
|
Source
|
Name of the Strain |
| Gauze |
- |
White |
White |
Seawater |
SA |
| GYP |
- |
White |
White |
Seawater |
Mar1 |
| GYP |
Dark |
White |
White-Gray |
Seawater |
Mar2 |
| ISP4 |
Black |
Yellow |
Red-Black |
Seawater |
R52 |
| SCA |
- |
White |
White |
Seawater |
Man1 |
| SCA |
- |
White |
White-Yellow |
Seawater |
Man2 |
| ISP2 |
White-Yellow |
White |
White |
Seawater |
SS |
| ISP2 |
Green- Black |
Brown |
Green-Gray |
Seawater |
SG |
| ISP2 |
A little dark |
Yellow |
White-Gray |
Seawater |
Cod1 |
| AIA |
- |
White |
White |
Seawater |
Cod2 |
| Gauze |
- |
White |
White-pink |
Soil |
GA1 |
| ISP4 |
- |
Violet |
White |
Soil |
GA2 |
| SCA |
- |
A little Yellow |
White |
Soil |
GA3 |
| ISP4 |
- |
White |
White-Pink |
Soil |
GA4 |
| ISP4 |
Black |
White |
White-Yellow |
Soil |
GA5 |
| Gauze |
Fire Brick |
White |
White |
Soil |
GA18 |
| Gauze |
Pink |
White |
White |
Soil |
GA20 |
| Gauze |
Dark green- Brown |
Dark |
White |
Soil |
GA31 |
| Gauze |
Blue- Purple |
A little dark |
White |
Soil |
GA33 |
| ISP2 |
Yellow |
Orange |
Orange |
Soil |
RZ |
| ISP4 |
Yellow |
Yellow |
White-Yellow |
Iris root |
EZ |
| Gauze |
- |
White |
White-Pink |
Echium Amoenum root |
EG |
Biological Assay of Crude Metabolite Extracts and Supernatant of the Isolated Strains
Anticancer Activity
The anti-cancer activity of crude extract and supernatant of the isolated strains was evaluated on the MCF-7 cell line (Figure 3) using MTT assay. The results showed that the supernatant of 19 isolates had an anticancer effect (Figure 4A). On the other hand, only the crude extract of 11 strains was able to inhibit MCF-7 cells growth (Figure 4B). According to the results, the supernatants showed higher anticancer activity than crude extracts. The highest growth inhibitory activity of bacterial supernatant belonged to ga31 (90.5%), followed by ez (82.3%), sa (76.9%), mar2 (70.2%), rz (65.7%), ga33 (56.5%), and ga5 (50.7%), respectively. Moreover, the highest growth inhibitory activity of crude extracts belonged to ga31 (75.5%), followed by ga5 (46.1%), rz (40.4%), and ez (40%), respectively. Surprisingly, we found a significant difference in the anticancer activity between the crude extract and supernatant (P < 0.05). Overall, we found that ga31 had the highest anticancer activity in both supernatant and crude extract form.
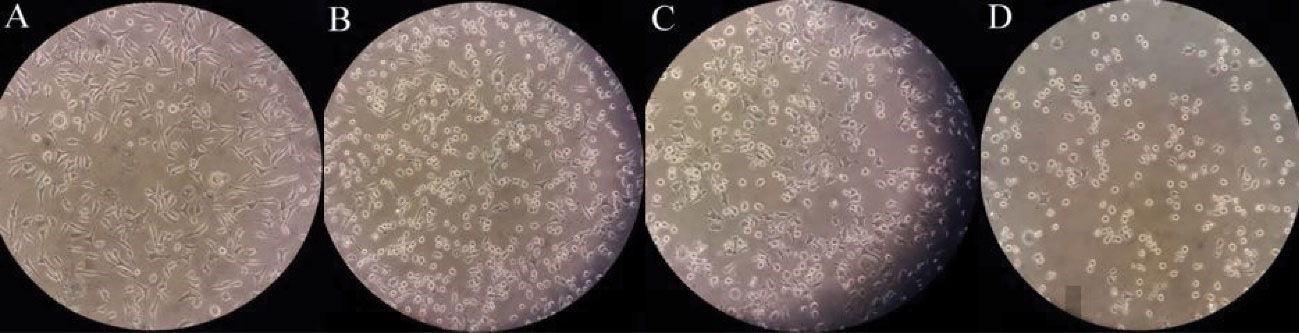
Figure 3.
Anticancer Activity of Isolated Actinobacteria Strains on McF-7, (A) Control, (B) ez, (C) sa, and (D) ga31
.
Anticancer Activity of Isolated Actinobacteria Strains on McF-7, (A) Control, (B) ez, (C) sa, and (D) ga31
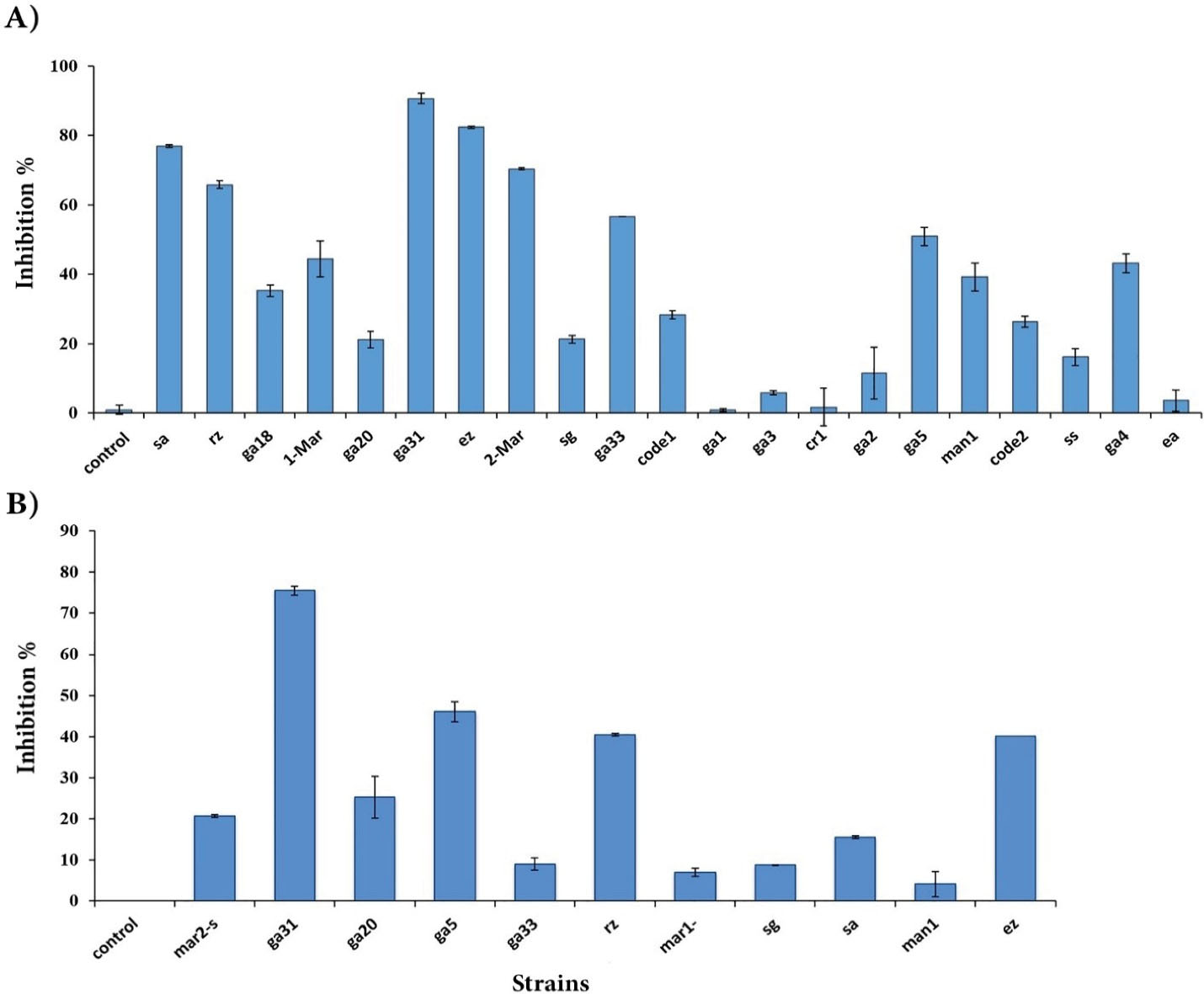
Figure 4.
Anticancer Effect of Supernatant (A), and Crude Extract (B) of Isolated Actinobacteria Strains on MCF-7 Cancer Cell Line. DMSO solvent (0.5%) was used in the control sample.
.
Anticancer Effect of Supernatant (A), and Crude Extract (B) of Isolated Actinobacteria Strains on MCF-7 Cancer Cell Line. DMSO solvent (0.5%) was used in the control sample.
Antibacterial Activity
The antibacterial activity of crude extract and supernatant of the isolated Actinobacteria strains was studied against two pathogenic bacteria, Pseudomonas aeruginosa PTCC 1214 and Staphylococcus aureus PTCC 1112, by disk diffusion method. The results showed that only the bacterial products of ga31 and ga5 strains had antibacterial activity against Staphylococcus aureus PTCC 1112 (Table 3).
Table 3.
Antibacterial Activity of Supernatant and Crude Extract of Two Strains (ga31 and ga5) on Staphylococcus Aureus PTCC 1112
|
Test
|
Inhibition Zone (mm)
|
MIC (µg/mL)
|
MBC (µg/mL)
|
|
Supernatant (Mean ± SD)
|
Crude Extract (Mean ± SD)
|
Supernatant
|
Crude Extract
|
Supernatant
|
Crude Extract
|
| Control* |
13.5±0.02 |
13.5±0.02 |
12.5 |
12.5 |
50 |
50 |
| ga31 |
14.8±0.08 |
16.3±0.19 |
15.6 |
7.8 |
62.5 |
31.25 |
| ga5 |
14.5±0.20 |
15.2±0.18 |
15.6 |
15.6 |
31.25 |
31.25 |
*Control: Gentamicin (10 µg) as a positive control in the disk diffusion method.
According to the results, Actinobacteria products (the supernatant and the crude extract) had not only bacteriostatic but also bactericidal activity against S. aureus PTCC 1112 (Table 3).
Identification of the Selected Strains
According to the results of biological tests, the ga31 strain had the highest anticancer and antimicrobial activities. Therefore, it was chosen for molecular identification. It was determined using the information obtained from the sequencing of the 16S rRNA gene. The results of the phylogenetic analysis of this strain by a maximum likelihood method in MEGA4 software show its evolutionary relationship with the Rubrobacteria class (Figure 5).
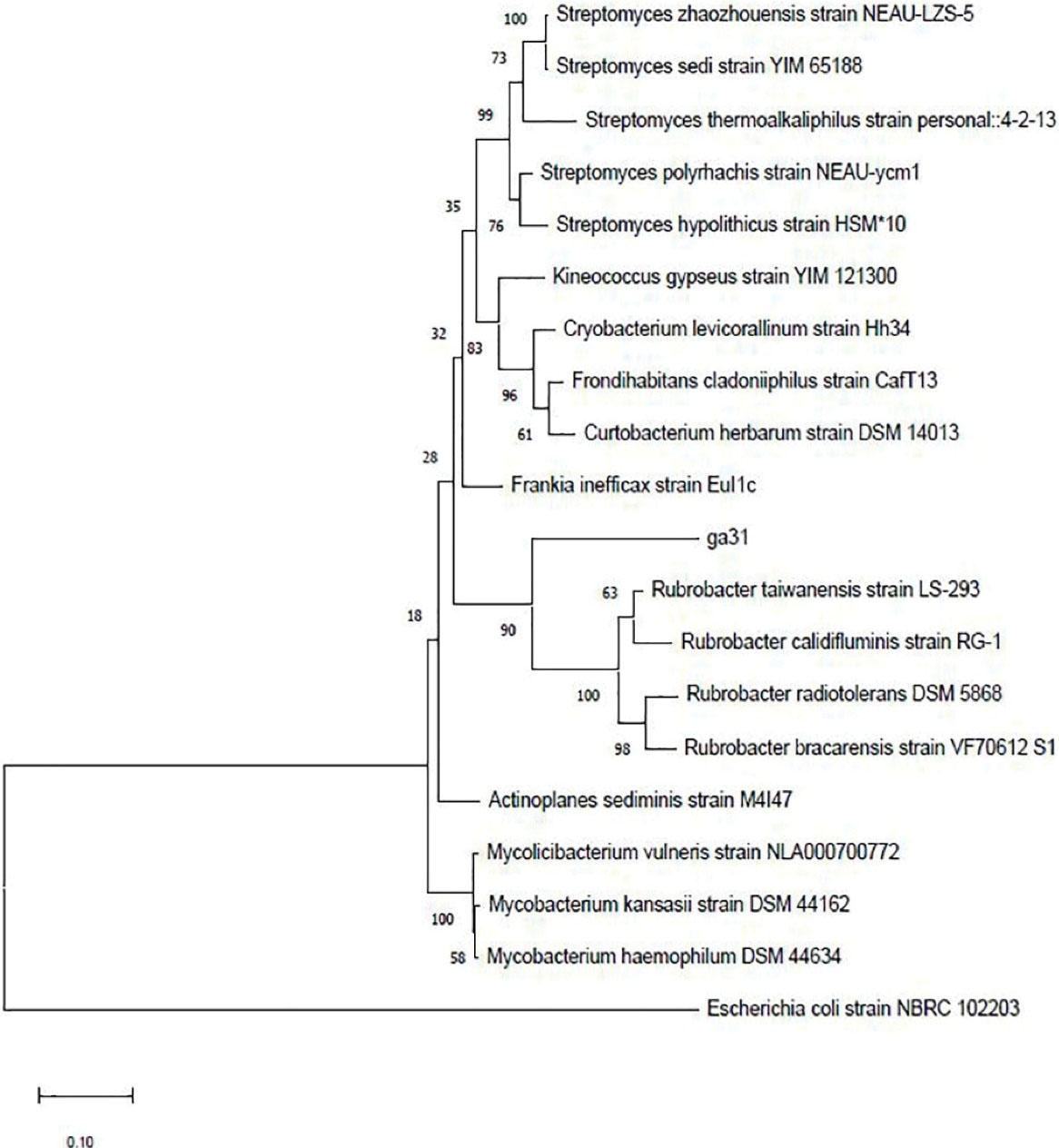
Figure 5.
The Phylogenetic Tree of ga31 Strain. The numbers at the nodes indicate the levels of bootstrap support based on 1000 resampled data sets. Escherichia coli Strain NBRC 102203 was used as an out-group.
.
The Phylogenetic Tree of ga31 Strain. The numbers at the nodes indicate the levels of bootstrap support based on 1000 resampled data sets. Escherichia coli Strain NBRC 102203 was used as an out-group.
Discussion
Actinobacteria are widespread in various ecosystems. These bacteria are important because of their ability to produce various secondary metabolites, including antibiotics, antifungals, anticancer, and immunosuppressive agents (5). Today, it is recommended that strains isolated from unusual and less studied habitats should be used to evaluate the bioactive and valuable compounds of these bacteria (25). Therefore, this study was conducted to investigate the biological potential of Actinobacteria isolated from various ecosystems of Iran, including the water of the Persian Gulf, the soil of the Gandom Beryan area in the Lut Desert, and the roots of some plants.
In this study, 112 Actinobacteria strains were isolated using morphological studies. Additionally, we investigated the strains by molecular tests using two specific primers of Actinobacteria (Sm6F, Sm5R) and (S-C-Act-235-a-S-20: F, S-C-Act-878-a-A-19: R) (26). The results confirmed that these isolated strains belonged to the Actinobacteria phylum. Accordingly, it was shown the wide distribution of different strains of Actinobacteria in ecosystems with different geographical conditions in this country. In the following, based on biochemical tests, 112 isolates were reduced to 22 strains whose anticancer and antimicrobial activities were examined.
As a result, we found that the supernatants of 7 isolated strains (ga31, ez, sa, mar2, rz, ga33, and ga5) had anticancer activity against the MCF-7 cell line. They inhibited the growth of the cancer cells by more than 50%. However, crude extracts of only 4 strains (ga31, ga5, rz, and ez) had anti-cancer activity. Based on morphological studies, the MCF-7 cell line became rounder and smaller cells after treatment with crude extracts of strains ez, sa, and ga31 (27). However, a significant difference in the anti-cancer activity was observed between crude extract and the supernatant of strains examined. It seems that this difference may be due to the dose-dependent activity of compounds produced (28) or the presence of compounds such as enzymes in the supernatant that have an anticancer effect but are not present in the crude extract. Nevertheless, among all the isolated strains, only the ga31 strain showed the maximum anti-cancer effect on the MCF-7 cells. Its supernatant and metabolite extract could inhibit the cancer cells by 90.5% and 75.5%, respectively. According to some evidence, different strains of Actinobacteria that were isolated from different sources had anticancer activity. They are able to inhibit the growth of various cancer cells. Azman et al reported that of the three novel rare actinobacteria strains isolated from mangrove soils at Tanjung Lumpur, Peninsular Malaysia, crude extract of two strains had anti-cancer effects against human cervical carcinoma cell line (Ca Ski). Ser HL et al in a study examined the anti-cancer effect of the Streptomyces pluripotens MUSC 137 strain isolated from mangrove soil. The results showed that MCF-7 cells were most susceptible to the extract with the lowest IC50 (61.33 ± 17.10 μg/mL), followed by HCT-116 and A549. Additionally, the results obtained from this study show the potential for the production of anti-cancer compounds by isolated strains (21,29-31).
Based on studies, living in diverse ecological conditions and Interaction with the environment and other organisms is a very important factor, because it can cause the chemical diversity of metabolites by this group of bacteria (6).
On the other hand, we found that of all isolated strains, only two strains (ga31 and ga5) had antimicrobial action against Staphylococcus aureus. However, they had no antimicrobial effect on Pseudomonas aeruginosa. The results obtained from the antimicrobial performance of these two strains are consistent with the results of previous studies. In a study, Rateb et al isolated and identified 3 new compounds from a Streptomyces strain. They observed that all three compounds had strong antibacterial activity against Gram-positive bacteria, but weak activity against Gram-negative bacteria. In the study conducted by Ding et al, of 53 actinomycetes strains isolated from desert ecosystems located in the northeast of Qinghai-Tibet Plateau, 26 strains were chosen for further studies, and according to the results, the metabolites produced mainly inhibited Gram-positive strains. According to reports, Actinobacteria isolated from desert environments have a better antimicrobial effect against gram-positive bacteria (32-35). It appears that the cell wall of Gram-negative bacteria has a key role in this function (32).
In general, according to the results obtained in this study, ga31 strain had both anti-cancer and antimicrobial effects. This strain belongs to the Actinobacteria phylum, isolated from the soil sample of Gandom Beryan area in the Lut Desert. The Lut Desert is one of the main deserts of the Iranian plateau and the twenty-fifth largest desert in the world. On the other hand, this region (Gandom Beryan) is known as the hottest place on earth (reaching a temperature of 70.7°C in summer). Therefore, the ga31 strain could be a good candidate for future studies to purify and evaluate new metabolites, and it is suggested that the performance of these compounds should be evaluated in in vivo studies.
Conclusions
Actinobacteria are very important producers of bioactive secondary metabolites. Our findings revealed that the products of isolated strains have biological effects and extreme ecosystems can be a source for the discovery of new bioactive compounds. Therefore, further research can be done to purify these compounds and use them in future studies.
Acknowledgments
This work was supported by the Shahid Bahonar University of Kerman. The authors are thankful to the management of this institution for providing necessary research facilities.
Conflict of Interests
The authors declare that they have no conflict of interests.
Ethical Approval
This article contains no studies with human participants or animals performed by any of the authors.
References
- Wei W, Zhou Y, Chen F, Yan X, Lai Y, Wei C. Isolation, diversity, and antimicrobial and immunomodulatory activities of endophytic actinobacteria from tea cultivars Zijuan and Yunkang-10 (Camellia sinensis var assamica). Front Microbiol 2018; 9:1304. doi: 10.3389/fmicb.2018.01304 [Crossref] [ Google Scholar]
- Govindasamy V, Franco CM, Gupta VV. Endophytic actinobacteria: diversity and ecology. In: Verma V, Gange A, eds. Advances in Endophytic Research. New Delhi: Springer; 2014. p. 27-59. 10.1007/978-81-322-1575-2_2.
- Lee LH, Zainal N, Azman AS, Eng SK, Goh BH, Yin WF. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. ScientificWorldJournal 2014; 2014:698178. doi: 10.1155/2014/698178 [Crossref] [ Google Scholar]
- Takahashi Y, Nakashima T. Actinomycetes, an inexhaustible source of naturally occurring antibiotics. Antibiotics (Basel) 2018; 7(2):45. doi: 10.3390/antibiotics7020045 [Crossref] [ Google Scholar]
- Davies-Bolorunduro OF, Adeleye IA, Akinleye MO, Wang PG. Anticancer potential of metabolic compounds from marine actinomycetes isolated from Lagos Lagoon sediment. J Pharm Anal 2019; 9(3):201-8. doi: 10.1016/j.jpha.2019.03.004 [Crossref] [ Google Scholar]
- van der Meij A, Worsley SF, Hutchings MI, van Wezel GP. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev 2017; 41(3):392-416. doi: 10.1093/femsre/fux005 [Crossref] [ Google Scholar]
- Barka EA, Vatsa P, Sanchez L, Gaveau-Vaillant N, Jacquard C, Meier-Kolthoff JP. Taxonomy, physiology, and natural products of actinobacteria. Microbiol Mol Biol Rev 2016; 80(1):1-43. doi: 10.1128/mmbr.00019-15 [Crossref] [ Google Scholar]
- Golinska P, Wypij M, Agarkar G, Rathod D, Dahm H, Rai M. Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Antonie Van Leeuwenhoek 2015; 108(2):267-89. doi: 10.1007/s10482-015-0502-7 [Crossref] [ Google Scholar]
- Manivasagan P, Venkatesan J, Sivakumar K, Kim SK. Pharmaceutically active secondary metabolites of marine actinobacteria. Microbiol Res 2014; 169(4):262-78. doi: 10.1016/j.micres.2013.07.014 [Crossref] [ Google Scholar]
- Suela Silva M, Naves Sales A, Teixeira Magalhães-Guedes K, Ribeiro Dias D, Schwan RF. Brazilian Cerrado soil actinobacteria ecology. Biomed Res Int 2013; 2013:503805. doi: 10.1155/2013/503805 [Crossref] [ Google Scholar]
- Nafis A, Raklami A, Bechtaoui N, El Khalloufi F, El Alaoui A, Glick BR. Actinobacteria from extreme niches in Morocco and their plant growth-promoting potentials. Diversity 2019; 11(8):139. doi: 10.3390/d11080139 [Crossref] [ Google Scholar]
- Ranjan R, Jadeja V. Isolation, characterization and chromatography based purification of antibacterial compound isolated from rare endophytic actinomycetes Micrococcus yunnanensis. J Pharm Anal 2017; 7(5):343-7. doi: 10.1016/j.jpha.2017.05.001 [Crossref] [ Google Scholar]
- Charousová I, Medo J, Hleba L, Císarová M, Javoreková S. Antimicrobial activity of actinomycetes and characterization of actinomycin-producing strain KRG-1 isolated from Karoo, South Africa. Braz J Pharm Sci 2019; 55:e17249. doi: 10.1590/s2175-97902019000217249 [Crossref] [ Google Scholar]
- Dhananjeyan V, Selvan N, Dhanapal K. Isolation, characterization, screening and antibiotic sensitivity of actinomycetes from locally (near MCAS) collected soil samples. J Biol Sci 2010; 10(6):514-9. doi: 10.3923/jbs.2010.514.519 [Crossref] [ Google Scholar]
- Stach JE, Maldonado LA, Ward AC, Goodfellow M, Bull AT. New primers for the class actinobacteria: application to marine and terrestrial environments. Environ Microbiol 2003; 5(10):828-41. doi: 10.1046/j.1462-2920.2003.00483.x [Crossref] [ Google Scholar]
- Yu J, Zhang L, Liu Q, Qi X, Ji Y, Kim BS. Isolation and characterization of actinobacteria from Yalujiang coastal wetland, North China. Asian Pac J Trop Biomed 2015; 5(7):555-60. doi: 10.1016/j.apjtb.2015.04.007 [Crossref] [ Google Scholar]
- Bruce KD, Hiorns WD, Hobman JL, Osborn AM, Strike P, Ritchie DA. Amplification of DNA from native populations of soil bacteria by using the polymerase chain reaction. Appl Environ Microbiol 1992; 58(10):3413-6. doi: 10.1128/aem.58.10.3413-3416.1992 [Crossref] [ Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 40. Mol Biol Evol 2007; 24(8):1596-9. doi: 10.1093/molbev/msm092 [Crossref] [ Google Scholar]
- Girão M, Ribeiro I, Ribeiro T, Azevedo IC, Pereira F, Urbatzka R. Actinobacteria isolated from Laminaria ochroleuca: a source of new bioactive compounds. Front Microbiol 2019; 10:683. doi: 10.3389/fmicb.2019.00683 [Crossref] [ Google Scholar]
- Manivasagan P, Gnanam S, Sivakumar K, Thangaradjou T, Vijayalakshmi S, Balasubramanian T. Antimicrobial and cytotoxic activities of an actinobacteria (Streptomyces sp PM-32) isolated from an offshore sediments of the Bay of Bengal in Tamilnadu. Adv Biol Res 2009; 3(5-6):231-6. [ Google Scholar]
- Azman AS, Othman I, Fang CM, Chan KG, Goh BH, Lee LH. Antibacterial, anticancer and neuroprotective activities of rare actinobacteria from mangrove forest soils. Indian J Microbiol 2017; 57(2):177-87. doi: 10.1007/s12088-016-0627-z [Crossref] [ Google Scholar]
- Senthilraja P, Kathiresan K. In vitro cytotoxicity MTT assay in Vero, HepG2 and MCF-7 cell lines study of Marine Yeast. J Appl Pharm Sci 2015; 5(3):80-4. doi: 10.7324/japs.2015.50313 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). M100-S24: Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement Wayne, PA: CLSI; 2014.
- Weseler A, Geiss HK, Saller R, Reichling J. A novel colorimetric broth microdilution method to determine the minimum inhibitory concentration (MIC) of antibiotics and essential oils against Helicobacter pylori. Pharmazie 2005; 60(7):498-502. [ Google Scholar]
- Mohamed H, Miloud B, Zohra F, García-Arenzana JM, Veloso A, Rodríguez-Couto S. Isolation and characterization of actinobacteria from Algerian Sahara soils with antimicrobial activities. Int J Mol Cell Med 2017; 6(2):109-20. doi: 10.22088/acadpub.BUMS.6.2.5 [Crossref] [ Google Scholar]
- Azadi D, Shojaei H, Mobasherizadeh S, Daei Naser A. Screening, isolation and molecular identification of biodegrading mycobacteria from Iranian ecosystems and analysis of their biodegradation activity. AMB Express 2017; 7(1):180. doi: 10.1186/s13568-017-0472-4 [Crossref] [ Google Scholar]
- Ser HL, Palanisamy UD, Yin WF, Chan KG, Goh BH, Lee LH. Streptomyces malaysiense sp nov: a novel Malaysian mangrove soil actinobacterium with antioxidative activity and cytotoxic potential against human cancer cell lines. Sci Rep 2016; 6:24247. doi: 10.1038/srep24247 [Crossref] [ Google Scholar]
- Anibou M, Chait A, Zyad A, Taourirt M, Ouhdouch Y, Benherref A. Actinomycetes from Moroccan habitats: isolation and screening for cytotoxic activities. World J Microbiol Biotechnol 2008; 24(10):2019-25. doi: 10.1007/s11274-008-9705-7 [Crossref] [ Google Scholar]
- Fiedler HP, Bruntner C, Riedlinger J, Bull AT, Knutsen G, Goodfellow M. Proximicin A, B and C, novel aminofuran antibiotic and anticancer compounds isolated from marine strains of the actinomycete Verrucosispora. J Antibiot (Tokyo) 2008; 61(3):158-63. doi: 10.1038/ja.2008.125 [Crossref] [ Google Scholar]
- Ser HL, Ab Mutalib NS, Yin WF, Chan KG, Goh BH, Lee LH. Evaluation of antioxidative and cytotoxic activities of Streptomyces pluripotens MUSC 137 isolated from mangrove soil in Malaysia. Front Microbiol 2015; 6:1398. doi: 10.3389/fmicb.2015.01398 [Crossref] [ Google Scholar]
- El-Shatoury SA, El-Shenawy NS, Abd El-Salam IM. Antimicrobial, antitumor and in vivo cytotoxicity of actinomycetes inhabiting marine shellfish. World J Microbiol Biotechnol 2009; 25(9):1547-55. doi: 10.1007/s11274-009-0040-4 [Crossref] [ Google Scholar]
- Ouchari L, Boukeskasse A, Bouizgarne B, Ouhdouch Y. Antimicrobial potential of actinomycetes isolated from the unexplored hot Merzouga desert and their taxonomic diversity. Biol Open 2019; 8(2):bio035410. doi: 10.1242/bio.035410 [Crossref] [ Google Scholar]
- Tiwari K, Upadhyay DJ, Mösker E, Süssmuth R, Gupta RK. Culturable bioactive actinomycetes from the Great Indian Thar Desert. Ann Microbiol 2015; 65(4):1901-14. doi: 10.1007/s13213-014-1028-3 [Crossref] [ Google Scholar]
- Rateb ME, Houssen WE, Harrison WT, Deng H, Okoro CK, Asenjo JA. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J Nat Prod 2011; 74(9):1965-71. doi: 10.1021/np200470u [Crossref] [ Google Scholar]
- Ding D, Chen G, Wang B, Wang Q, Liu D, Peng M. Culturable actinomycetes from desert ecosystem in northeast of Qinghai-Tibet Plateau. Ann Microbiol 2013; 63(1):259-66. doi: 10.1007/s13213-012-0469-9 [Crossref] [ Google Scholar]