Avicenna Journal of Clinical Microbiology and Infection. 8(4):130-134.
doi: 10.34172/ajcmi.2021.24
Original Article
Prevalence of Metallo-β-Lactamase Genes in Clinical Isolates of Klebsiella pneumoniae in Health Care Centers in Mazandaran Province
Rougayeh Alizadeh 1, Majid Alipour 1, 2, *  , Fatereh Rezaee 1
, Fatereh Rezaee 1
Author information:
1Department of Cell and Molecular Biology, Babol Branch, Islamic Azad University, Babol, Iran
2Comprehensive Health Research Center, Babol Branch, Islamic Azad University, Babol, Iran
*
Corresponding author: Majid Alipour, PhD in Cellular and Molecular Biology, Assistant professor, Department of Cell and Molecular biology, Babol Branch, Islamic Azad University, Babol, Iran Email:
alipourmk@gmail.com
Abstract
Background:
Klebsiella pneumoniae is one of the most important causes of opportunistic infections, including lobar pneumonia, urinary tract infection (UTIs), and wound infection. Treatment of disease caused by this bacterium has also become a challenge, as many strains are resistant to all β-lactam antibiotics. Metallo-β-lactamases hydrolyze most β-lactam antibiotics, especially carbapenems, but cannot inactivate monobactams. This study aimed to determine the prevalence of Metallo-β-lactamase genes (blaIMP, blaVIM, and blaNDM) in K. pneumoniae isolated from clinical specimens.
Methods: Clinical samples were collected from hospitals of Mazandaran province. Among 500 clinical samples collected, only 40 Klebsiella pneumoniae isolates were detected by culture and biochemical tests. Antimicrobial susceptibility testing was performed on all isolates by the Kirby-Bauer method. The polymerase chain reaction (PCR) technique was used for the identification of blaIMP, blaVIM, and blaNDM genes.
Results: Antibiogram by disk diffusion method showed that 21 (52%) and 19 (48%) isolates were classified as imipenem resistant and sensitive, respectively. Of all the samples, 30 (75%), 7 (17.5%), and 36 (90%) contained blaVIM, blaIMP, and blaNDM genes, respectively. The co-existence of the blaVIM and blaNDM genes was observed in 22 (55%) isolates. The presence of both blaIMP and blaNDM genes was confirmed in 2 (5%) of the isolates. Four isolates (10%) had blaNDM, blaIMP, and blaVIM genes simultaneously, but none of these genes were present in one isolate (2.5%).
Conclusions: This study showed that the prevalence of metallo-β-lactamase genes in K. pneumoniae isolated from clinical specimens is very high, so it is recommended that physicians treat patients based on phenotypic and genotypic characteristics.
Keywords: Metallo-β-lactamase, Resistance gene, Klebsiella pneumoniae, Clinical samples
Copyright and License Information
© 2021 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Klebsiella pneumoniae is a clinically significant pathogen that causes urinary tract infections (UTIs), sepsis, pneumonia, and other infections (1). The bacterium is a Gram-negative, immobile, encapsulated, lactose-fermenting, and facultative anaerobic bacillus (2). Klebsiella pneumoniae can asymptomatically colonize the skin, mouth, respiratory tract, and gastrointestinal tract, which may serve as a reservoir for human infections (3). The bacterium uses a variety of strategies to protect itself against antibiotics, one of which is the production of β-lactamase enzymes. This enzyme causes resistance by hydrolyzing and inactivating the β-lactam ring of beta-lactam antibiotics. Treatment of disease caused by K. pneumoniae has become a challenge because many strains are resistant to all β-lactam antibiotics through the production of carbapenemases (4). According to the Ambler classification, beta-lactamases are grouped into classes A, B, C, and D based on sequence similarity (5). Class B beta-lactamases require at least one or two zinc ions at their active site (6). Various beta-lactamases, including Verona integron-encoded metallo-β-lactamase (VIM), imipenemase (IMP), New Dehli metallo-β-lactamase (NDM-1), Sao Paulo metallo β-lactamase (SPM), and Seol imipenemase metallo-β-lactamase (SIM), have been detected in K. pneumoniae strains (7). Metallo-β-lactamases cause resistance to various antibiotics, including penicillins, cephalosporins, and carbapenems, except for monobactam (8,9). IMP is a carbapenemase that is resistant to almost all β-lactam antibiotics. One of the most common types of IMP is IMP-4, which was first identified in Acinetobacter spp. in Hong Kong in 2001 (10). VIM metallo-β-lactamases were first identified in Europe (11). The first case of VIM-4 in K. pneumoniae was reported in a recent study conducted in Tunisia (12). In 2009, NDM was first detected in a Swedish patient hospitalized in New Delhi, India (9). The genes encoding these carbapenemases are reported to be located on chromosomes or motile genetic elements such as plasmids, integrons, and transposons (13). Based on several reports on the presence of Metallo-β-lactamases genes in K. pneumoniae strains, the aim of the present study was to determine the prevalence of blaIMP, blaVIM, and blaNDM genes in clinical isolates.
Methods
Isolation and Detection of Klebsiella pneumoniae
Clinical specimens were collected from wounds, burns, urine, and sputum. The study was conducted for seven months from June 2018 to December 2019. In total, 40 isolates of K. pneumoniae were transferred from the laboratories of Mazandaran province to the Microbiology Laboratory of Islamic Azad University, Babol branch. Isolates were inoculated on MacConkey agar plates and were incubated overnight at 37°C (Merck, Germany). Unique colonies of K. pneumoniae (lactose-fermenting mucoid colonies ) were confirmed by standard biochemical tests, including fermentation of sugars in Triple Sugar Iron Agar medium (TSI) (Merck, Germany), fermentation of acidic or alcoholic in MRVP (Methyl Red, Voges–Proskauer) broth (Merck, Germany), growth in Simmons citrate medium (Merck, Germany), urea hydrolysis in urea medium, production of indole and hydrogen sulfide, and motility in sulfide indole motility (SIM) medium (Merck, Germany). According to biochemical tests, K. pneumoniae isolates were confirmed as indole negative, MR negative, VP positive, citrate positive, urease positive, and motility negative (14). All K. pneumoniae isolates were cultured in trypticase soy broth (TSB) (Merck, Germany). Then, 800 μL of bacterial suspension and 200 μL of glycerol were mixed in a 1.5 mL tube and were stored at -20°C for further studies (15,16).
Antibiotic Susceptibility Test
According to the Clinical and Laboratory Standards Institute (CLSI) recommendation, antibiotic susceptibility testing was performed by the disk diffusion method. A bacterial suspension with turbidity equal to 0.5 McFarland was cultured on the surface of Müller-Hinton agar medium. Then, the imipenem disc (10 μg) (Padtan Teb Co-Iran) was placed on the culture medium and was incubated at 37°C for 18 hours. The diameter of the growth inhibition zone was measured and the results were recorded as resistant, intermediate, and sensitive. Isolates with growth inhibition zones of ≥22 mm, 20-22 mm, and ≤19 mm were determined as sensitive, intermediate, and resistant, respectively (17).
Detection of Metallo -β-Lactamase Genes
The polymerase chain reaction (PCR) technique was used for the identification of metallo-beta-lactamase genes including blaNDM, blaIMP, and blaVIM in K. pneumoniae isolates. DNA purification was performed by boiling method. The two colonies were transferred into a test tube containing 1 mL of distilled water, boiled in a water bath for 10 minutes, and centrifuged at 1000 rpm for 5 minutes. Then, 3 μL of supernatant was used in the PCR master mix (18). PCR amplification was performed in a final volume of 25 μL containing 12.5 μL of Master mix (CinnaGen Company, Iran), 3 μL of supernatant containing bacterial DNA, 1 μL of each primer (20 pmol), and 7.5 μL of sterile distilled water. All PCR assays were performed using an Eppendorf 5331 MasterCycler Gradient Thermal Cycler (Hamburg, Germany). The primers used for detection of these genes were listed in Table 1.
Table 1.
The Sequences of Primers Used to Identify NDM, IMP, and VIM Genes
|
Gene
|
Primers
5′→3′
|
Amplicon
Base Pair (bp)
|
Annealing
Temperature
|
References |
|
bla
NDM
|
F-ACCGCCTGGACCGATGACCA
R-GCCAAAGTTGGGCGCGGTTG |
236 |
69 ° C |
(18) |
|
bla
IMP
|
F- GAAGGCGTTTATGTTCATAC
R- GTATGTTTCAAGAGTGATGC |
587 |
52.3 ° C |
(18) |
|
bla
VIM
|
F- GTTTGGTCGCATATCGCAAC
R- AATGCGCAGCACCAGGATAG |
382 |
63 ° C |
(19) |
Thermal cycles of PCR were performed as follows. A cycle of initial denaturation was performed at 94°C for 3 minutes. Then, 35 cycles, including 94°C for 1 minute, annealing at a suitable temperature for 1 minute according to Table 1, and polymerization at 72°C for 1 minute were carried out. Final polymerization was done at 72°C for 10 minutes (19,20). In the PCR reaction mixture, distilled water was used as the negative control. Finally, PCR products were electrophoresed in 2% agarose gel at 95 V for 100 minutes and were observed by UV transilluminator (Vilber Lourmat, Collégien, France).
Statistical Analysis
The results of this study were analyzed using SPSS (Statistical Package for Social Sciences) version 21.0.
Results
A total of 40 K. pneumoniae isolates were identified from clinical specimens. K. pneumoniae strains were isolated from 11 sputum, 21 urine, 3 blood, and 5 wound samples. The results of antimicrobial susceptibility testing performed on 40 isolates of K. pneumoniae showed that 21 (52%) and 19 (48%) isolates were resistant and sensitive to imipenem, respectively, and none of the samples were intermediate (Figure 1).

Figure 1.
Antibiogram of Klebsiella Pneumoniae Strains to Imipenem by the Disk Diffusion Method R: resistant; S: sensitive
.
Antibiogram of Klebsiella Pneumoniae Strains to Imipenem by the Disk Diffusion Method R: resistant; S: sensitive
PCR technique was used to identify metallo-β-lactamase resistance genes. Agarose gel electrophoresis of PCR products produced special bands of 236, 587, and 383 bp for blaNDM, blaIMP, and blaVIM, respectively (Figures 2, 3, and 4).
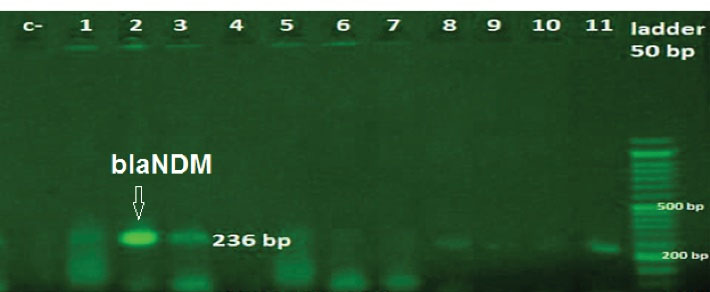
Figure 2.
Agarose Gel Electrophoresis of the PCR Amplicon of blaNDM. Ladder: (50 bp); Lanes 2,3,8 and 11: positive samples; Lanes c-: negative control; Lane 1, 4, 6, 7, 9, and 10: negative samples.
.
Agarose Gel Electrophoresis of the PCR Amplicon of blaNDM. Ladder: (50 bp); Lanes 2,3,8 and 11: positive samples; Lanes c-: negative control; Lane 1, 4, 6, 7, 9, and 10: negative samples.
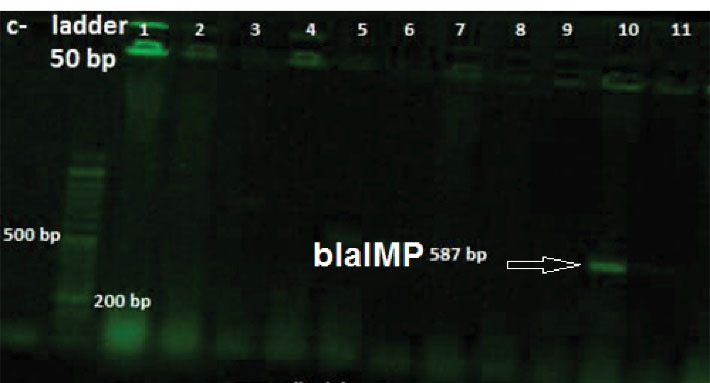
Figure 3.
Agarose Gel Electrophoresis of the PCR Amplicon of blaIMP. Ladder: (50 bp); Lanes 10: positive samples; Other lanes: negative samples; c-: negative control.
.
Agarose Gel Electrophoresis of the PCR Amplicon of blaIMP. Ladder: (50 bp); Lanes 10: positive samples; Other lanes: negative samples; c-: negative control.
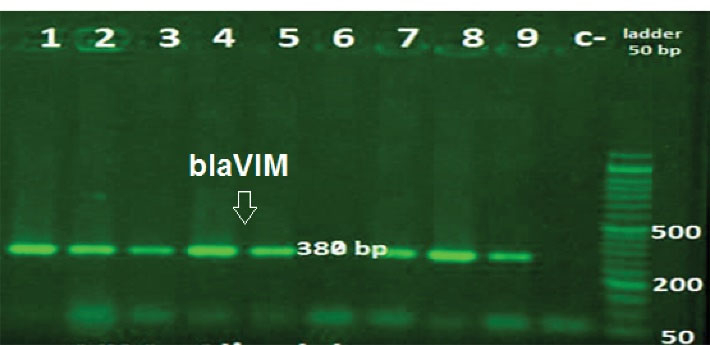
Figure 4.
Agarose Gel Electrophoresis of the PCR Amplicon of blaVIM. Ladder: (50 bp); Other lanes: positive samples; Lanes 6: negative samples; c-: negative control.
.
Agarose Gel Electrophoresis of the PCR Amplicon of blaVIM. Ladder: (50 bp); Other lanes: positive samples; Lanes 6: negative samples; c-: negative control.
The prevalence of metallo-β-lactamase genes is shown in Table 2.
Table 2.
Frequency of Metallo-β-Lactamase Resistance Genes
|
Gene
|
Number (%)
|
|
bla
NDM
|
36 (90) |
|
bla
IMP
|
7 (17.5) |
|
bla
VIM
|
30 (75) |
|
bla
NDM+
bla
IMP
|
2 (5) |
|
bla
NDM+
bla
VIM
|
22 (55) |
|
bla
IMP+
bla
VIM
|
0 (0) |
|
bla
NDM+
bla
IMP+
bla
VIM
|
4 (10) |
| None |
1 (2.5) |
| Total |
40 (100) |
Of the 40 K. pneumoniae isolates studied, the most common resistance gene was the blaNDM gene with a frequency of 36 (90%), but the blaIMP gene had the lowest prevalence (17.5%). The prevalence of the blaVIM gene was 75%. The coexistence of the blaNDM, blaIMP, and blaVIM was observed in 4 (10%) isolates of K. pneumoniae. The coexistence of blaNDM andblaIMP as well as blaNDM andblaVIM genes was detected in 2 (5%) and 22 (55%) isolates, respectively. The presence of both blaIMP andblaVIM genes was not detected in any of the isolates. Moreover, 2.5% of the isolates lacked beta-lactamase resistance genes.
Discussion
Klebsiella pneumoniae plays a clinically significant role in hospitalized infections. The bacterium causes many infections, such as UTIs, liver abscesses, pneumonia, and bacteremia (21). In our study, blaNDM gene was detected in 90% of the K. pneumoniae isolates, which differs from studies conducted in Northwest Pakistan (22.7%) (22), India (19%) (23), China (14.9%) (24), Egypt (26.32%) (25), and Bangladesh (22.4%) (26). In a study conducted by Saremi et al in Iran, the prevalence of blaNDM-1 gene was 1.92% (7). In a study conducted in Peru, 85% of Klebsiella pneumoniae isolateswere positive for blaNDM gene, which is consistent with our study (27). The prevalence of the blaNDM gene varies in different parts of the world depending on the level of health and overuse of antibiotics. In the current study, the blaIMP gene was observed in 17.5% of the isolates, which is not consistent with studies conducted in South India (8%) (28), China (36.5%) (29), and Egypt (15.2%) (30). In the present study, the prevalence of the blaVIM gene was 75%. In a study conducted by Ripabelli et al (31), blaVIM gene was detected in 76.5% of the isolates, which is consistent with our study. In our study, the coexistence of blaNDMandblaIMP as well as blaNDMandblaVIM genes was observed in 5% and 55% of the isolates, respectively. In a study conducted by Urmi et al (26), multiple Metallo-beta-lactamase genes were present simultaneously in one K. pneumoniae isolate. The results of a study conducted by Hussein showed the coexistence of both blaNDM-1 and blaIMP genes in only three strains of K. pneumoniae (32). These studies show that the simultaneous presence of resistance genes varies in different geographical areas. In our study, 1 (2.5%) isolate did not contain any of the metallo-beta-lactamase resistance genes. In an investigation carried out by Pragasam et al (33), 4 isolates were negative for all the genes tested, which is consistent with our study. In the genotyping method, 97% of K. pneumoniae isolates contained Metallo-beta-lactamase resistance genes, but in the phenotyping method, 52% of isolates showed resistance to imipenem. Different phenotyping and genotyping results can be due to mutations in the promoter or coding sequence of metallo-beta-lactamase genes, which in turn prevent gene transcription and enzyme inefficiency, respectively. It is recommended that physicians use the results of this study to select the appropriate antibiotics to treat patients. Carbapenem-resistant K. pneumoniae strains are usually resistant to most beta-lactam antibiotics. Therefore, treatment options are limited to polymyxins, tigecycline, and aminoglycosides (34).
Conclusions
Due to the high prevalence of metallo-β-lactamase genes in this study and their roles in treatment failure, rapid detection of these strains in laboratories plays a more influential role in controlling the spread of these strains. Therefore, appropriate policies are needed to regulate the use of penicillins, cephalosporins, and carbapenems in patients. We recommend regular monitoring and screening for the emergence of K. pneumoniae strains containing the metallo-beta-lactamase genes.
Acknowledgments
The authors would like to thank Molecular Biology Laboratory of Islamic Azad University, Babol Branch.
Authors’ Contribution
Majid Alipour designed the study, analyzed data, wrote the manuscript, and carried out the interpretation of the results. Rougayeh Alizadehand Fatereh Rezaee performed experimental tests and prepared the samples. All authors read and approved the final manuscript.
Conflict of Interests
There is no conflict of interest for authors.
Ethical Approval
There are no ethical issues for this article.
Funding/Support
None.
References
- Yamasaki S, Shigemura K, Osawa K, Kitagawa K, Ishii A, Kuntaman K. Genetic analysis of ESBL-producing Klebsiella pneumoniae isolated from UTI patients in Indonesia. J Infect Chemother 2021; 27(1):55-61. doi: 10.1016/j.jiac.2020.08.007 [Crossref] [ Google Scholar]
- Alizadeh Behbahani B, Tabatabaei Yazdi F, Shahidi F, Noorbakhsh H, Vasiee A, Alghooneh A. Phytochemical analysis and antibacterial activities extracts of mangrove leaf against the growth of some pathogenic bacteria. Microb Pathog 2018; 114:225-32. doi: 10.1016/j.micpath.2017.12.004 [Crossref] [ Google Scholar]
- Chen CM, Wang M, Li XP, Li PL, Tian JJ, Zhang K. Homology analysis between clinically isolated extraintestinal and enteral Klebsiella pneumoniae among neonates. BMC Microbiol 2021; 21(1):25. doi: 10.1186/s12866-020-02073-2 [Crossref] [ Google Scholar]
- Palmieri M, D’Andrea MM, Pelegrin AC, Mirande C, Brkic S, Cirkovic I. Genomic epidemiology of carbapenem- and colistin-resistant Klebsiella pneumoniae isolates from Serbia: predominance of ST101 strains carrying a novel OXA-48 plasmid. Front Microbiol 2020; 11:294. doi: 10.3389/fmicb.2020.00294 [Crossref] [ Google Scholar]
- Bae DW, Jung YE, Jeong BG, Cha SS. Novel inhibition mechanism of carbapenems on the ACC-1 class C β-lactamase. Arch Biochem Biophys 2020; 693:108570. doi: 10.1016/j.abb.2020.108570 [Crossref] [ Google Scholar]
- Grewal AS, Thapa K, Sharma N, Singh S. New Delhi metallo-β-lactamase-1 inhibitors for combating antibiotic drug resistance: recent developments. Med Chem Res 2020; 29(8):1301-20. doi: 10.1007/s00044-020-02580-x [Crossref] [ Google Scholar]
- Saremi M, Saremi L, Feizy F, Vafaei S, Lashkari A, Saltanatpour Z. The prevalence of VIM, IMP, and NDM-1 metallo-beta-lactamase genes in clinical isolates of Klebsiella pneumoniae in Qom province, Iran. J Med Microbiol Infect Dis 2020; 8(1):34-9. doi: 10.29252/JoMMID.8.1.34 [Crossref] [ Google Scholar]
- Khaledi M, Shahini Shams Abadi M, Validi M, Zamanzad B, Vafapour R, Gholipour A. Phenotypic and genotypic detection of metallo-β-lactamases in A baumanii isolates obtained from clinical samples in Shahrekord, southwest Iran. BMC Res Notes 2019; 12(1):597. doi: 10.1186/s13104-019-4636-y [Crossref] [ Google Scholar]
- Kalasseril SG, Krishnan R, Vattiringal RK, Paul R, Mathew P, Pillai D. Detection of New Delhi metallo-β-lactamase 1 and cephalosporin resistance genes among carbapenem-resistant Enterobacteriaceae in water bodies adjacent to hospitals in India. Curr Microbiol 2020; 77(10):2886-95. doi: 10.1007/s00284-020-02107-y [Crossref] [ Google Scholar]
- Xu J, Lin W, Chen Y, He F. Characterization of an IMP-4-producing Klebsiella pneumoniae ST1873 strain recovered from an infant with a bloodstream infection in China. Infect Drug Resist 2020; 13:773-9. doi: 10.2147/idr.s247341 [Crossref] [ Google Scholar]
- Luzzaro F, Docquier JD, Colinon C, Endimiani A, Lombardi G, Amicosante G. Emergence in Klebsiella pneumoniae and Enterobacter cloacae clinical isolates of the VIM-4 metallo-beta-lactamase encoded by a conjugative plasmid. Antimicrob Agents Chemother 2004; 48(2):648-50. doi: 10.1128/aac.48.2.648-650.2004 [Crossref] [ Google Scholar]
- Tayh G, Nagarjuna D, Sallem RB, Verma V, Chairat S, Boudabous A. First report of VIM metallo-β-lactamase production in Escherichia coli and Klebsiella pneumoniae clinical isolates from Gaza Strip, Palestine. Germs 2020; 10(1):18-26. doi: 10.18683/germs.2020.1181 [Crossref] [ Google Scholar]
- Taggar G, Attiq Rheman M, Boerlin P, Diarra MS. Molecular epidemiology of carbapenemases in Enterobacteriales from humans, animals, food and the environment. Antibiotics (Basel) 2020; 9(10):693. doi: 10.3390/antibiotics9100693 [Crossref] [ Google Scholar]
- Firoozeh F, Mahluji Z, Khorshidi A, Zibaei M. Molecular characterization of class 1, 2 and 3 integrons in clinical multi-drug resistant Klebsiella pneumoniae isolates. Antimicrob Resist Infect Control 2019; 8:59. doi: 10.1186/s13756-019-0509-3 [Crossref] [ Google Scholar]
- Qian W, Wang W, Zhang J, Wang T, Liu M, Yang M. Antimicrobial and antibiofilm activities of ursolic acid against carbapenem-resistant Klebsiella pneumoniae. J Antibiot (Tokyo) 2020; 73(6):382-91. doi: 10.1038/s41429-020-0285-6 [Crossref] [ Google Scholar]
- Alipour M, Mirabbasi R, Azizi IG, Yahyapour Y. Isolation and detection of Vibrio vulnificus from coastal seawater of Babolsar. Zahedan J Res Med Sci 2018; 20(7):e67004. doi: 10.5812/zjrms.67004 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). M100: Performance Standards for Antimicrobial Susceptability Testing. 30th ed. Wayne, PA: CLSI; 2020.
- Dashti AA, Jadaon MM, Abdulsamad AM, Dashti HM. Heat treatment of bacteria: a simple method of DNA extraction for molecular techniques. Kuwait Med J 2009; 41(2):117-22. [ Google Scholar]
- Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, Nasiri S, Owlia P, Nikbin VS. First report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb Drug Resist 2013; 19(1):30-6. doi: 10.1089/mdr.2012.0078 [Crossref] [ Google Scholar]
- Shams S, Hashemi A, Esmkhani M, Kermani S, Shams E, Piccirillo A. Imipenem resistance in clinical Escherichia coli from Qom, Iran. BMC Res Notes 2018; 11(1):314. doi: 10.1186/s13104-018-3406-6 [Crossref] [ Google Scholar]
- Chen T, Dong G, Zhang S, Zhang X, Zhao Y, Cao J. Effects of iron on the growth, biofilm formation and virulence of Klebsiella pneumoniae causing liver abscess. BMC Microbiol 2020; 20(1):36. doi: 10.1186/s12866-020-01727-5 [Crossref] [ Google Scholar]
- Khaliq A, Rahman H, Khaliq H, Khan Z, Khan TA, Sheer R. blaKPC, blaNDM and blaOXA-48 Producing Klebsiella pneumoniae: First Report from Northwest Pakistan. J Pharmacy Pharmacol 2021; 9:1-9. [ Google Scholar]
- Veeraraghavan B, Shankar C, Karunasree S, Kumari S, Ravi R, Ralph R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog Glob Health 2017; 111(5):240-6. doi: 10.1080/20477724.2017.1340128 [Crossref] [ Google Scholar]
- Kong Z, Liu X, Li C, Cheng S, Xu F, Gu B. Clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Jiangsu province, China. Infect Drug Resist 2020; 13:4627-35. doi: 10.2147/idr.s293206 [Crossref] [ Google Scholar]
- El-Badawy MF, El-Far SW, Althobaiti SS, Abou-Elazm FI, Shohayeb MM. The first Egyptian report showing the co-existence of blaNDM-25, blaOXA-23, blaOXA-181, and blaGES-1 among carbapenem-resistant K pneumoniae clinical isolates genotyped by BOX-PCR. Infect Drug Resist 2020; 13:1237-50. doi: 10.2147/idr.s244064 [Crossref] [ Google Scholar]
- Urmi UL, Nahar S, Rana M, Sultana F, Jahan N, Hossain B. Genotypic to phenotypic resistance discrepancies identified involving β-lactamase genes, blaKPC, blaIMP, blaNDM-1, and blaVIM in uropathogenic Klebsiella pneumoniae. Infect Drug Resist 2020; 13:2863-75. doi: 10.2147/idr.s262493 [Crossref] [ Google Scholar]
- Roach D, Waalkes A, Abanto J, Zunt J, Cucho C, Soria J. Whole genome sequencing of Peruvian Klebsiella pneumoniae identifies novel plasmid vectors bearing carbapenem resistance gene NDM-1. Open Forum Infect Dis 2020; 7(8):ofaa266. doi: 10.1093/ofid/ofaa266 [Crossref] [ Google Scholar]
- Indrajith S, Mukhopadhyay AK, Chowdhury G, Farraj DAA, Alkufeidy RM, Natesan S. Molecular insights of carbapenem resistance Klebsiella pneumoniae isolates with focus on multidrug resistance from clinical samples. J Infect Public Health 2021; 14(1):131-8. doi: 10.1016/j.jiph.2020.09.018 [Crossref] [ Google Scholar]
- Dong F, Zhang Y, Yao K, Lu J, Guo L, Lyu S. Epidemiology of carbapenem-resistant Klebsiella pneumoniae bloodstream infections in a Chinese children’s hospital: predominance of New Delhi metallo-β-lactamase-1. Microb Drug Resist 2018; 24(2):154-60. doi: 10.1089/mdr.2017.0031 [Crossref] [ Google Scholar]
- Moemen D, Masallat DT. Prevalence and characterization of carbapenem-resistant Klebsiella pneumoniae isolated from intensive care units of Mansoura University Hospitals. Egypt J Basic Appl Sci 2017; 4(1):37-41. doi: 10.1016/j.ejbas.2017.01.001 [Crossref] [ Google Scholar]
- Ripabelli G, Tamburro M, Guerrizio G, Fanelli I, Flocco R, Scutellà M. Tracking multidrug-resistant Klebsiella pneumoniae from an Italian hospital: molecular epidemiology and surveillance by PFGE, RAPD and PCR-based resistance genes prevalence. Curr Microbiol 2018; 75(8):977-87. doi: 10.1007/s00284-018-1475-3 [Crossref] [ Google Scholar]
- Hussein NH. Emergence of NDM-1 among carbapenem-resistant Klebsiella pneumoniae in Iraqi hospitals. Acta Microbiol Immunol Hung 2018; 65(2):211-27. doi: 10.1556/030.64.2017.026 [Crossref] [ Google Scholar]
- Pragasam AK, Shankar C, Anandan S, Verghese VP, Veeraraghavan B. Alarming increase in carbapenemase-producing Klebsiella spp causing bloodstream infections in pediatric population in India. J Infect Dev Ctries 2017; 11(9):736-7. doi: 10.3855/jidc.8038 [Crossref] [ Google Scholar]
- Vivas R, Dolabella SS, Barbosa AAT, Jain S. Prevalence of Klebsiella pneumoniae carbapenemase - and New Delhi metallo-beta-lactamase-positive K pneumoniae in Sergipe, Brazil, and combination therapy as a potential treatment option. Rev Soc Bras Med Trop 2020; 53:e20200064. doi: 10.1590/0037-8682-0064-2020 [Crossref] [ Google Scholar]