Avicenna Journal of Clinical Microbiology and Infection. 7(3):75-80.
doi: 10.34172/ajcmi.2020.17
Original Article
Molecular Detection of Arcobacter in Human Stool Samples Using Housekeeping Genes
Aysan Karamghoshchi 1  , Azam Ahmadi 1, *
, Azam Ahmadi 1, *  , Mohammad Arjomandzadegan 1, Majid Akbari 1, Elahe Ghorbani Marghmaleki 1
, Mohammad Arjomandzadegan 1, Majid Akbari 1, Elahe Ghorbani Marghmaleki 1
Author information:
1Department of Microbiology, Infectious Diseases Research Center (IDRC), Arak University of Medical Sciences, Arak, Iran.
Abstract
Background:
Arcobacter is one of the most common bacteria in humans and livestock, leading to gastroenteritis in humans as well as genital and enteric diseases in animals. This bacterium is known to be the main cause of diarrhea. In molecular studies, the 16SrRNA gene was primarily used as the standard gene for the determination of the Arcobacter. The purpose of this study was to investigate the molecular detection of Arcobacter using glyA, atpA, and gyrA genes compared to16SrRNA.
Methods: In this study, 61 samples of Arcobacter DNA isolated from fecal specimens of patients and healthy individuals in the sample bank were used. In order to detect Arcobacter, the intended primers for 16SrRNA as well as glyA, atpA, and gyrA genes were used for polymerase chain reaction (PCR). The products obtained from the PCR were sequenced.
Results: The results of the proliferation reactions indicated the accuracy of the intended primers and the associated molecular experiments. Our results showed that 65.57% of the cases were detected to be positive for Arcobacter among 61 samples using the glyA gene. This percentage was higher compared to 16SrRNA (42.62%), gyrA (42.62%), and atpA (24.59%). The analysis was statistically significant.
Conclusions: Given the presence of repetitive sequences in the 16SrRNA in most bacteria, the interpretation of the results is likely to be difficult for researchers. The results of this study showed more sensitivity and accurate diagnosis of Arcobacter using the glyA gene than other studied genes. In diagnostic studies of Arcobacter, the glyA gene is proposed as an alternative to the 16SrRNA.
Keywords: Arcobacter, Polymerase chain reaction, Diarrhea, 16SrRNA, glyA
Copyright and License Information
© 2020 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Arcobacter, Campylobacter, and Helicobacter are members of the Campylobacter family (1). In 1978, Arcobacter spp. were first isolated from aborted bovine fetuses in England. To date, 22 species have been identified, including Arcobacter butzleri, Arcobacter cryaerophilus, and so on. They are clinically important and are related to human and pathogens of animals and have also been found in stools of people with diarrhea and samples of individuals with bacteremia, endocarditis, and peritonitis (2). A number of studies have also reported the isolation of Arcobacter from stool in healthy humans (3). A. butzleri is most commonly associated with human disease including enteritis or watery diarrhea (3,4). According to previous studies, in different countries such as South Africa, Belgium, and France, A. butzleri is one of the important pathogens in human stools (5,6). Arcobacter is the cause of persistent diarrhea (7,8). Arcobacter spp. have also been isolated from different biological samples of various animals (9-11). This bacterium can be transmitted through products of animal origin, water, and even oral-fecal contact with humans (12,13). The livestock and poultry carcasses can be contaminated with water, feces, and viscera infected with Arcobacter in slaughterhouses (14). This bacterium can be contaminated by contact with raw meat and even by eating unclean meat (15,16). Vegetables may also be contaminated if they are washed with contaminated water or in contact with contaminated feces (17). Animals are, therefore, significant reservoirs of Arcobacter spp. Although different methods and environments have been used to distinguish Arcobacter from different samples, a standard and accurate reference method has not been provided. Phenotypic and differential microbiological methods for confirmation of Arcobacter colonization, including oxidase, catalase, nitrate regeneration, hydrophilic hydrolysis, acetate indoxyl hydrolysis, growth in air and at 25ºC, growth in 4% salt, growth in MacConkey medium, and resistance to cephoperazone (18). Due to phenotypic properties of Arcobacter, its detection by molecular methods is more accurate and realistic (19). Several molecular diagnostic methods have been developed to improve sensitivity and reduce the time needed to detect Arcobacter (20). However, molecular techniques are often difficult and costly. To date, no study has been conducted on accurate molecular detection of Arcobacter spp. using the glyA, atpA, and gyrA genes simultaneously.
By targeting 16SrRNA and 23SrRNA genes, one-step PCR can be used to detect A. butzleri, A. cryaerophilus, and A. skirrowi at the same time (21). Other molecular methods have been proposed, including real-time PCR, DGGE, and AFLP (22). 16SrRNA gene is significantly conserved within the species of the genus so that it can be used as a golden standard for bacterial diagnosis. Although 16SrRNA gene is an indicator gene and the design of the primers is easy to amplify, it has many variables for the differentiation of microbial taxa. Almost all the studies in this field have been done on the molecular determination of Arcobacter using this gene. However, there are also some disadvantages. The 16SrRNA nucleotide sequence has multiple copies and low molecular resolution and cannot be easily interpreted in a research framework (23,24). In this way, the use of other housekeeping genes such as gyrA (encoding DNA gyrase submit A), glyA (encoding serine transhydroxymethylase), and atpA (encoding the submission of F1 ATPase) may offer different potential benefits for the molecular detection of Arcobacter (25).
The objective of this study is to identify Arcobacter in human stool samples using other housekeeping genes, including gyrA, glyA, and atpA, in order to develop target genes in addition to the 16SrRNA gene for molecular detection.
Materials and Methods
Collection of Samples
In this study, we selected 61 genomic DNA samples extracted from human fecal specimens available from the DNA Bank of Infectious Diseases Research Center (Arak University of Medical Sciences). These DNA samples were extracted from stool samples and were enriched with special media (arco broth) containing antibiotics incubated at 28°C for 48 hours. Then, they were inoculated on Brucella agar medium followed by passive filtration of the broth through a 0.45 μm membrane filter placed on the blood agar medium (26). Among these DNA samples, 29 samples were from healthy people who were exposed to poultry meat, and 32 samples were from individuals with diarrhea.
Genus-Specific PCR
Genus-specific PCR has been used to detect Arcobacter at the level of the genus. Primers for gyrA, glyA, and atpA genes have been designed using specialized programs such as Primer Blast, Mega 4.0, Oligo 6.0, and Primer3. The sequences of 16SrRNA-specific primers were provided by Gonzalez et al (15). The specifications of the used primers are shown in Table 1. The PCR reaction mix for each gene included 1.5 μL of extracted DNA (20-50 μg), 0.7 μL of each primer (Copenhagen, Denmark), 7.5 μL of 2x super master-mix (YTA, Iran), and 4.6 μL of DDW in the final volume of 15 μL. The amplification was performed with initial denaturation at 94°C (5 minutes) followed by 28 cycles of denaturation at 94°C (1 minute), annealing at specific temperatures (55 seconds), and extension at 72°C (55 seconds) (Table 1). Moreover, the final extension was carried out at 72°C (8 minutes). DNA extracted from the Arcobacter colonies and the water (no template) were considered positive and negative, respectively.
Table 1.
Specifications of the Primers Used in the Study
|
Primer
|
Sequences (5 to 3)
|
Target Genes
|
Size of Products (bp)
|
Annealing
°C
|
Arc1
Arc2 |
AGAACGGGTTATAGCTTGCTAT
GATACAATACAGGCTAATCTCT |
16SrRNA |
181 |
52.7 |
GyrA F
GyrA R |
GAGATCAAGGAAGAAGTACAAG
TGTATTTCTTCCTGCTTTTCTAATTG |
gyrA
|
330 |
52.7 |
GlyA F
GlyA R |
AGCAGCTAATGAACATCCAAGT
CCACCTTGAAGTCCTGGGAA |
glyA
|
175 |
52.7 |
AtpA F
AtpA R |
TCAAGCTGGAGACGTTGC
ATTGTGCAAACGCCTCAAGT |
atpA
|
220 |
65 |
Gel Electrophoresis
Gel electrophoresis was used to evaluate the PCR results of each amplifier. The products of PCR reactions were loaded on a 1.3% agarose gel (Genefanavaran, Iran). The results were analyzed using the gel doc system (Quantum ST4, Germany).
Sequencing
PCR products were sequenced on an ABI automatic sequencer (Applied Biosystem Inc., CA, USA) using Macrogen (South Korea) facilities for confirmation of the amplification reaction.
Statistical Analysis
Statistical analyses were carried out using related software packages such as Excel 2007 and MedCalc 18.11.
Results
Evaluation of PCR Products for Electrophoresis
Figure 1 shows the PCR products electrophoresed on 1.3% agarose gel.
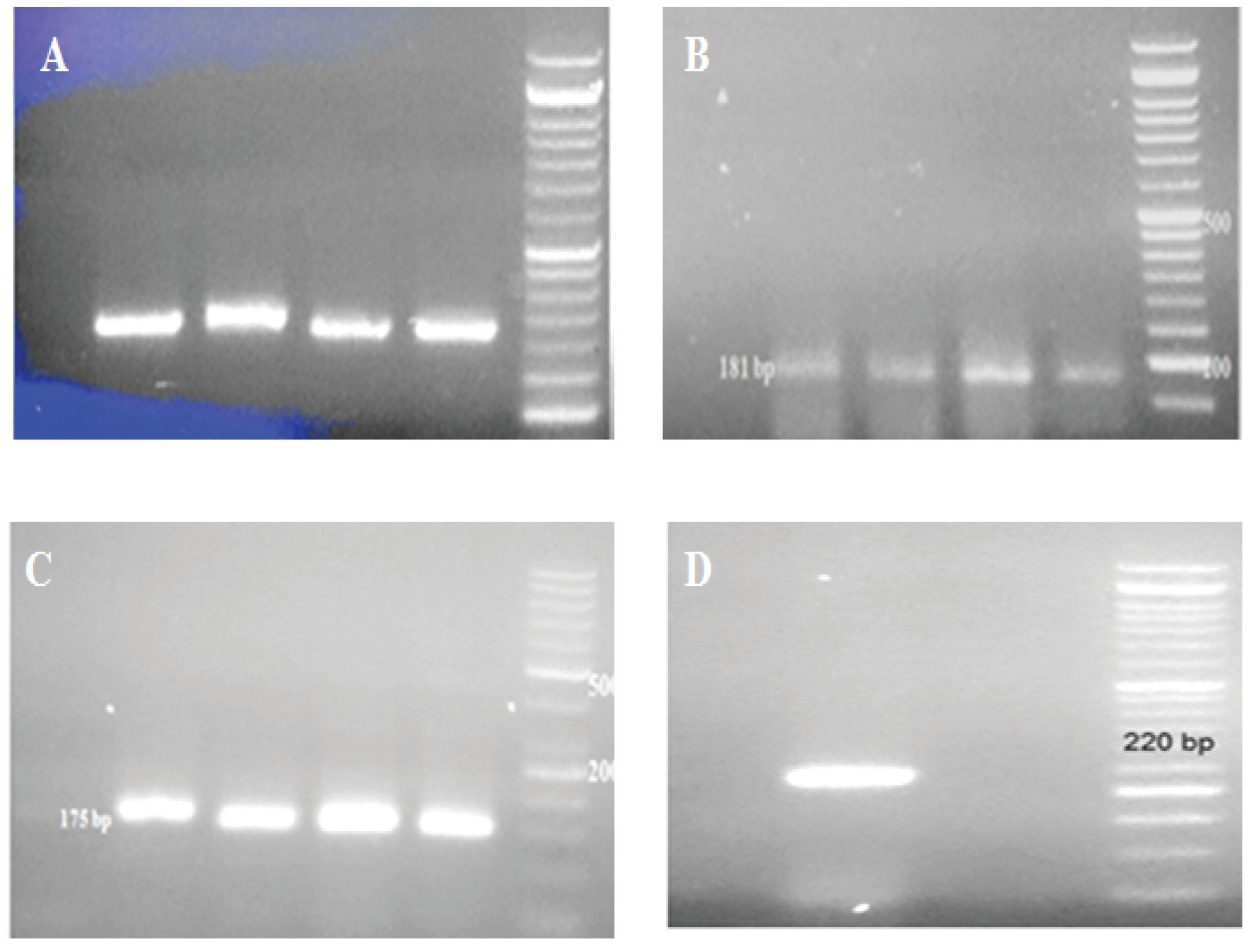
Figure 1.
Electrophoresis of PCR Products of gyrA (A), 16SrRNA (B), glyA (C), and atpA (D) on 1.3% Agarose Gel. Product size: 330 bp, 181 bp, 175 bp, and 220 bp, respectively. L lane: ladder with size of 50 bp (YTA Co.). C- Lane: negative control. C+ Lane: positive control.
.
Electrophoresis of PCR Products of gyrA (A), 16SrRNA (B), glyA (C), and atpA (D) on 1.3% Agarose Gel. Product size: 330 bp, 181 bp, 175 bp, and 220 bp, respectively. L lane: ladder with size of 50 bp (YTA Co.). C- Lane: negative control. C+ Lane: positive control.
The sequencing results were analyzed using related software packages (Mega4 and Chromas). The amplicons confirmed the data (Figure 2).
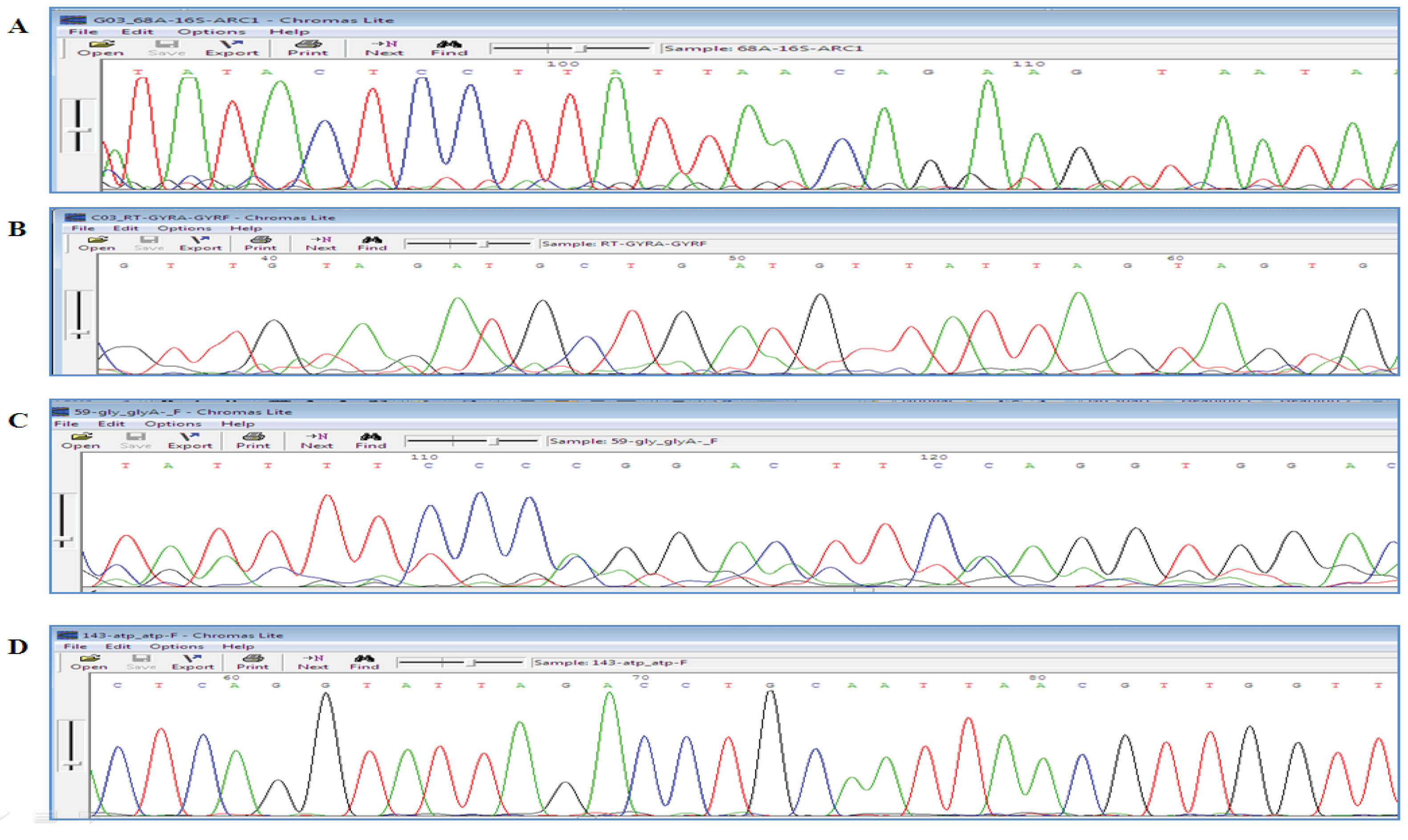
Figure 2.
The Results of Sequencing of Genes: (A) 16SrRNA (max score: 186, QC: 80%, Percent Identity: 91.34%, E value: 1e-46), B) gyrA, (max score: 223, QC: 89%, Percent Identity: 95.45%, E value: 4e-58), (C) glyA, (max score: 196, QC: 53%, Percent Identity: 95.20%, E value: 8e-50), (D) atpA, (max score: 322, QC: 96%, Percent Identity: 96.95%, E value: 8e-88).
.
The Results of Sequencing of Genes: (A) 16SrRNA (max score: 186, QC: 80%, Percent Identity: 91.34%, E value: 1e-46), B) gyrA, (max score: 223, QC: 89%, Percent Identity: 95.45%, E value: 4e-58), (C) glyA, (max score: 196, QC: 53%, Percent Identity: 95.20%, E value: 8e-50), (D) atpA, (max score: 322, QC: 96%, Percent Identity: 96.95%, E value: 8e-88).
PCR Results
Of the 61 samples studied, 26 cases (42.62%) of 16SrRNA gene PCR, 26 cases (42.62%) of gyrA gene PCR, 40 cases (65.57%) of glyA gene PCR, and 15 cases (24.59%) of atpA gene PCR were positive for Arcobacter, as detailed in Table 2.
Table 2.
Frequency of Positive Samples in Molecular Detection by Each Gene
|
atpA
|
gyrA
|
16SrRNA
|
glyA
|
Samples
|
| 9 (14.75%) |
9 (14.75%) |
9 (14.75%) |
19 (31.14%) |
Healthy people |
| 6 (9.83%) |
17 (27.86%) |
17 (27.86%) |
21 (34.42%) |
Patient |
Discussion
The increased isolation of Arcobacter from clinical samples and healthy people has increased its importance in general health (27). Due to the existence of Arcobacter in food products containing animal resources, vegetables, and water, this bacterium is introduced as a food-borne pathogen (28). In addition, Arcobacter can cause gastroenteritis in human and genital diseases in animals, which can confirm the need to detect Arcobacter (29). Because molecular detection methods are faster and more accurate than cultural ones, we used molecular methods to identify Arcobacter in fecal samples of healthy individuals and patients. In previous studies, the 16SrRNA gene is mostly used for the diagnosis of the Arcobacter gene. Due to the existence of different bacterial genomes in stool samples as well as repeated nucleotide sequences in the 16SrRNA gene of some bacteria, the detection of Arcobacter probably cannot be accurate (25). Therefore, in this study, we evaluated the molecular detection of Arcobacter using housekeeping genes such as gyrA, glyA, and atpA genes compared to 16SrRNA genes. Based on the suggestions of other studies, these genes have been selected. To date, no study has been conducted in our study environment reporting similar prevalence rates for Arcobacter using these genes. To the best of our knowledge, no previous study has used four housekeeping genes (gyrA, glyA, atpA, and 16SrRNA) simultaneously in the stool samples to differentiate Arcobacter species. Luis Collado et al proposed the use of the gyrA gene in the PCR-hybridation method (30). William G Miller et al reported the application of the glyA gene for genotyping Arcobacter spp. using the MLST technique (4). In a study conducted by Miller et al, the PCR-RFLP technique was used for proliferation and sequencing of atpA to differentiate Campylobacteraceae and helicobacteraceae families (31). Al Rashid et al detected Campylobacter jejuni, C. coli, C. lari, upsaliensis, Arcobacter butzleri species by the glyA gene. In their study, a PCR-hybridization method was developed in which primers are used to amplify glyA fragments. Evaluation of this strategy with genomic DNA from different strains has shown that the above-mentioned method is specific and sensitive (32). Therefore, in the genes used in our study, glyA was also selected. Abdelbaqi et al studied the development of real-time PCR for investigating the quinolone resistance-determining regions in the gyrA gene of Arcobacter spp. in France (33). According to their study, the nucleotide sequences of the gyrA genes of A. butzleri, A. cryaerophilus, A. cibarius, and A. skirrowi were determined. Phylogenetic analysis of gyrA sequences provides results similar to phylogenetic analysis of the 16SrRNA gene sequence and allows for differentiation between A. butzleri species (33).
In our study, in addition to the 16SrRNA gene, we used other in-house genes ( gyrA, glyA, and atpA ) to detect Arcobacter at the molecular level. As shown in Table 2, 19 (31.14%), 9 (14.75%), 9 (14.75%), 9 (14.75%) of healthy people, who were exposed to poultry meat, and 21 (34.42%), 17 (27.86%), 17 (27.86%), 6 (9.83%) of patients with diarrheawere detected positive using proliferation of glyA, 16SrRNA, gyrA, and atpA genes, respectively, among a total of 61 samples. Recently, it has been shown that Arcobacter can be better identified by glyA gene than other genes in both groups. Although this bacterium has been detected more frequently in the patient group than in healthy people, glyA may be useful for the identification of Arcobacter in both groups (P=0.01). In addition, according to statistical data, the sensitivity of the use of the glyA gene to detect the Arcobacter is higher compared to the 16SrRNA gene. As shown in Table 3, the difference in the prevalence between groups was statistically significant (P < 0.05). Therefore, the proliferation of glyA gene by designing correct primers, which can be attached to genomes of different species of the Arcobacter, may be more useful than other studied genes for the detection of Arcobacter. In this study, due to limited financial resources, we did not examine all the housekeeping genes. We used only 4 genes for screening, and in future studies, other housekeeping genes can also be used. Despite the above discussion, it is recommended that this study should be carried out with a larger sample size and that the bacterial load of Arcobacter be studied in fecal samples of both healthy people and patients.
Table 3.
Statistical Value of glyA Gene Compared With 16SrRNA
|
Diagnosis Analyses
|
n=61
|
Percent
|
Group
|
Result
|
Gene
|
Sensitivity: 52.830%
(38.636-66.68%) |
32 Patients
29 Healthy people |
27.86 |
17 Patients |
26 Positive |
Identification by 16SrRNA |
| 14.75 |
9 Healthy people |
| Specificity: 68.750% (53.749-81.340% |
24.59 |
15 Patients |
35 Negative |
| 32.78 |
20 Healthy people |
| Sensitivity: 66.038% (51.733-78.480) |
32 Patients
29 Healthy people |
34.42 |
21 Patients |
40 Positive |
Identification by glyA |
| 31.14 |
19 Healthy people |
Specificity: 34.043%
(20.864-49.31%) |
18.03 |
11 Patients |
21 Negatives |
| 16.39 |
10 Healthy people |
Chi-squared (trend), 5.754; DF, 1; Significance level, P =.0165.
Conclusions
To date, Arcobacter has not been detected using gyrA, atpA, and glyA genes in clinical samples. The results of this study have shown that the glyA gene is more acceptable than other used housekeeping genes for molecular detection of this bacteria. Proliferation of the glyA gene may be considered as an alternative to the 16SrRNA gene to detect Arcobacter genus.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgement
The authors would like to thank the research assistant and all co-workers for helping us to represent this study.
Ethical Approval
This study was derivate from research project of Arak University of medical science with IR.ARAKMU.REC.1397.228 ethical code.
Authors’ Contribution
AK: performed the experiments and manuscript preparation; AA: designed study, interpreted data, manuscript preparation and approved final manuscript; MA: designed study and interpretation of data; MA: clinical sample preparation and analyzed data; EGM: performed the experiments and interpretation of data
Funding/Support
This work was supported under Grant Number 3255 by Arak University of Medical Sciences.
References
- Collins CI, Wesley IV, Murano EA. Detection of Arcobacter spp in ground pork by modified plating methods. J Food Prot 1996; 59(5):448-52. doi: 10.4315/0362-028x-59.5.448 [Crossref] [ Google Scholar]
- Webb AL, Taboada EN, Selinger LB, Boras VF, Inglis GD. Efficacy of wastewater treatment on Arcobacter butzleri density and strain diversity. Water Res 2016; 105:291-6. doi: 10.1016/j.watres.2016.09.003 [Crossref] [ Google Scholar]
- Houf K, Stephan R. Isolation and characterization of the emerging foodborn pathogen Arcobacter from human stool. J Microbiol Methods 2007; 68(2):408-13. doi: 10.1016/j.mimet.2006.09.020 [Crossref] [ Google Scholar]
- Wesley IV, Miller WG. Arcobacter: an opportunistic human foodborne pathogen? In: Scheld WM, Grayson ML, Hughes JM, eds. Emerging Infections 9. Washington, DC: American Society of Microbiology Press; 2010. p. 185-211.
- Engberg J, On SL, Harrington CS, Gerner-Smidt P. Prevalence of Campylobacter, Arcobacter, Helicobacter, and Sutterella spp in human fecal samples as estimated by a reevaluation of isolation methods for campylobacters. J Clin Microbiol 2000; 38(1):286-91. [ Google Scholar]
- Van den Abeele AM, Vogelaers D, Van Hende J, Houf K. Prevalence of Arcobacter species among humans, Belgium, 2008-2013. Emerg Infect Dis 2014; 20(10):1731-4. doi: 10.3201/eid2010.140433 [Crossref] [ Google Scholar]
- Ferreira S, Queiroz JA, Oleastro M, Domingues FC. Genotypic and phenotypic features of Arcobacter butzleri pathogenicity. Microb Pathog 2014; 76:19-25. doi: 10.1016/j.micpath.2014.09.004 [Crossref] [ Google Scholar]
- On SL, Stacey A, Smyth J. Isolation of Arcobacter butzleri from a neonate with bacteraemia. J Infect 1995; 31(3):225-7. doi: 10.1016/s0163-4453(95)80031-x [Crossref] [ Google Scholar]
- Shirzad Aski H, Tabatabaei M, Khoshbakht R, Raeisi M. Occurrence and antimicrobial resistance of emergent Arcobacter spp isolated from cattle and sheep in Iran. Comp Immunol Microbiol Infect Dis 2016; 44:37-40. doi: 10.1016/j.cimid.2015.12.002 [Crossref] [ Google Scholar]
- Andersen MM, Wesley IV, Nestor E, Trampel DW. Prevalence of Arcobacter species in market-weight commercial turkeys. Antonie Van Leeuwenhoek 2007; 92(3):309-17. doi: 10.1007/s10482-007-9153-7 [Crossref] [ Google Scholar]
- Ellström P, Hansson I, Söderström C, Engvall EO, Rautelin H. A prospective follow-up study on transmission of Campylobacter from poultry to abattoir workers. Foodborne Pathog Dis 2014; 11(9):684-8. doi: 10.1089/fpd.2014.1753 [Crossref] [ Google Scholar]
- Patyal A, Rathore RS, Mohan HV, Dhama K, Kumar A. Prevalence of Arcobacter spp in humans, animals and foods of animal origin including sea food from India. Transbound Emerg Dis 2011; 58(5):402-10. doi: 10.1111/j.1865-1682.2011.01221.x [Crossref] [ Google Scholar]
- Fernandez H, Villanueva MP, Mansilla I, Gonzalez M, Latif F. Arcobacter butzleri and A cryaerophilus in human, animals and food sources, in southern Chile. Braz J Microbiol 2015; 46(1):145-7. doi: 10.1590/s1517-838246120140095 [Crossref] [ Google Scholar]
- Khoshbakht R, Tabatabaei M, Shirzad Aski H, Seifi S. Occurrence of Arcobacter in Iranian poultry and slaughterhouse samples implicates contamination by processing equipment and procedures. Br Poult Sci 2014; 55(6):732-6. doi: 10.1080/00071668.2014.971223 [Crossref] [ Google Scholar]
- González I, García T, Antolín A, Hernández PE, Martín R. Development of a combined PCR-culture technique for the rapid detection of Arcobacter spp in chicken meat. Lett Appl Microbiol 2000; 30(3):207-12. doi: 10.1046/j.1472-765x.2000.00696.x [Crossref] [ Google Scholar]
- Kabeya H, Maruyama S, Morita Y, Ohsuga T, Ozawa S, Kobayashi Y. Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int J Food Microbiol 2004; 90(3):303-8. doi: 10.1016/s0168-1605(03)00322-2 [Crossref] [ Google Scholar]
- Vandamme P, Vancanneyt M, Pot B, Mels L, Hoste B, Dewettinck D. Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb nov and Arcobacter skirrowii sp nov, an aerotolerant bacterium isolated from veterinary specimens. Int J Syst Bacteriol 1992; 42(3):344-56. doi: 10.1099/00207713-42-3-344 [Crossref] [ Google Scholar]
- Houf K, On SLW, Coenye T, Mast J, Van Hoof J, Vandamme P. Arcobacter cibarius sp nov, isolated from broiler carcasses. Int J Syst Evol Microbiol 2005; 55(Pt 2):713-7. doi: 10.1099/ijs.0.63103-0 [Crossref] [ Google Scholar]
- On SL. Identification methods for campylobacters, helicobacters, and related organisms. Clin Microbiol Rev 1996; 9(3):405-22. doi: 10.1128/cmr.9.3.405-422.1996 [Crossref] [ Google Scholar]
- Petersen RF, Harrington CS, Kortegaard HE, On SL. A PCR-DGGE method for detection and identification of Campylobacter, Helicobacter, Arcobacter and related Epsilobacteria and its application to saliva samples from humans and domestic pets. J Appl Microbiol 2007; 103(6):2601-15. doi: 10.1111/j.1365-2672.2007.03515.x [Crossref] [ Google Scholar]
- Houf K, Tutenel A, De Zutter L, Van Hoof J, Vandamme P. Development of a multiplex PCR assay for the simultaneous detection and identification of Arcobacter butzleri, Arcobacter cryaerophilus and Arcobacter skirrowii. FEMS Microbiol Lett 2000; 193(1):89-94. doi: 10.1111/j.1574-6968.2000.tb09407.x [Crossref] [ Google Scholar]
- Quiñones B, Parker CT, Janda JM, Jr Jr, Miller WG, Mandrell RE. Detection and genotyping of Arcobacter and Campylobacter isolates from retail chicken samples by use of DNA oligonucleotide arrays. Appl Environ Microbiol 2007; 73(11):3645-55. doi: 10.1128/aem.02984-06 [Crossref] [ Google Scholar]
- Walsh DA, Bapteste E, Kamekura M, Doolittle WF. Evolution of the RNA polymerase B’ subunit gene (rpoB’) in Halobacteriales: a complementary molecular marker to the SSU rRNA gene. Mol Biol Evol 2004; 21(12):2340-51. doi: 10.1093/molbev/msh248 [Crossref] [ Google Scholar]
- Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol 2010; 76(12):3886-97. doi: 10.1128/aem.02953-09 [Crossref] [ Google Scholar]
- Fox GE, Wisotzkey JD, Jurtshuk P Jr. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 1992; 42(1):166-70. doi: 10.1099/00207713-42-1-166 [Crossref] [ Google Scholar]
- Collado L, Figueras MJ. Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev 2011; 24(1):174-92. doi: 10.1128/cmr.00034-10 [Crossref] [ Google Scholar]
- Hausdorf L, Neumann M, Bergmann I, Sobiella K, Mundt K, Fröhling A. Occurrence and genetic diversity of Arcobacter spp in a spinach-processing plant and evaluation of two Arcobacter-specific quantitative PCR assays. Syst Appl Microbiol 2013; 36(4):235-43. doi: 10.1016/j.syapm.2013.02.003 [Crossref] [ Google Scholar]
- Banting GS, Braithwaite S, Scott C, Kim J, Jeon B, Ashbolt N. Evaluation of various Campylobacter-specific quantitative PCR (qPCR) assays for detection and enumeration of Campylobacteraceae in irrigation water and wastewater via a miniaturized most-probable-number–qPCR assay. Appl Environ Microbiol 2016; 82(15):4743-56. doi: 10.1128/aem.00077-16 [Crossref] [ Google Scholar]
- Neill SD, Campbell JN, O’brien JJ, Weatherup ST, Ellis WA. Taxonomic position of Campylobacter cryaerophila sp nov. Int J Syst Evol Microbiol 1985; 35(3):342-56. doi: 10.1099/00207713-35-3-342 [Crossref] [ Google Scholar]
- Collado L, Cleenwerck I, Van Trappen S, De Vos P, Figueras MJ. Arcobacter mytili sp nov, an indoxyl acetate-hydrolysis-negative bacterium isolated from mussels. Int J Syst Evol Microbiol 2009; 59(Pt 6):1391-6. doi: 10.1099/ijs.0.003749-0 [Crossref] [ Google Scholar]
- Miller WG, Yee E, Jolley KA, Chapman MH. Use of an improved atpA amplification and sequencing method to identify members of the Campylobacteraceae and Helicobacteraceae. Lett Appl Microbiol 2014; 58(6):582-90. doi: 10.1111/lam.12228 [Crossref] [ Google Scholar]
- Al Rashid ST, Dakuna I, Louie H, Ng D, Vandamme P, Johnson W. Identification of Campylobacter jejuni, C coli, C lari, C upsaliensis, Arcobacter butzleri, and A butzleri-like species based on the glyA gene. J Clin Microbiol 2000; 38(4):1488-94. doi: 10.1128/jcm.38.4.1488-1494.2000 [Crossref] [ Google Scholar]
- Abdelbaqi K, Ménard A, Prouzet-Mauleon V, Bringaud F, Lehours P, Mégraud F. Nucleotide sequence of the gyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol Med Microbiol 2007; 49(3):337-45. doi: 10.1111/j.1574-695X.2006.00208.x [Crossref] [ Google Scholar]