Avicenna Journal of Clinical Microbiology and Infection. 7(2):40-44.
doi: 10.34172/ajcmi.2020.09
Original Article
Frequency and Antimicrobial Susceptibility of Multidrug-resistant Klebsiella pneumoniae Isolated From Wound Samples in Isfahan, Iran
Fahimeh Nourbakhsh 1  , Samaneh Borooni 2
, Samaneh Borooni 2  , Elaheh Tajbakhsh 3, *
, Elaheh Tajbakhsh 3, *  , Dana Daneshmand 4
, Dana Daneshmand 4
Author information:
1Medical Toxicology Research Centre, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran.
2Nourdanesh Institutes of Higher Education, Meymeh, Isfahan, Iran.
3Departments of Microbiology, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran.
4Nosocomial Infection Research Center, Isfahan University of Medical Science, Isfahan, Iran.
*
Corresponding author: Elaheh Tajbakhsh, Department of Microbiology, Faculty of Basic Sciences, Shahrekord Branch, Islamic Azad University, Iran. Email:
ee_tajbakhsh@yahoo.com
Abstract
Background: Klebsiella pneumoniae is one of the most important opportunistic enteric bacteria and is a major cause of pneumonia and urinary tract infection. In addition, the serotype capsules of K1 and K2 can cause intense diseases. Further, the acquisition of plasmid that codes the production of extended-spectrum β-lactamases (ESBLs) confers K. pneumoniae resistance on a number of broad-spectrum antibiotics posing a global public health problem. Accordingly, this study aimed to identify 120 K. pneumoniae isolates that were detected from infected wound samples in Isfahan hospitals in Iran.
Methods: Capsular serotypes and antibiotic resistance genes were studied in 120 isolates of K. pneumoniae from different clinical cases in Isfahan, Iran. To this end, the frequency of resistance genes at the presence of specific primers was examined and all resistant isolates were tested for the detection of capsular serotypes genes using special primers.
Results: The results demonstrated that 120 isolates had serotype K2 with the redundancy of 78% and most cases had serotype K5 with the redundancy of 63%. Based on the results, aac (3)-IV gene was observed in most isolates with the redundancy of 54.1% and tetA with the redundancy of 75.86%. In this study, the highest resistance belonged to ceftazidime (74.3%), ciprofloxacin (78.5%), and tetracycline (72). Furthermore, the results revealed that serotype K2 is one of the most important serotypes of K. pneumonia. Finally, there seems to be a strong relationship between the presence of integron and increased resistance to different antibiotics.
Conclusions: In general, this was the first extensive study regarding the distribution and antimicrobial resistant profile of K. pneumoniae and related genes. Therefore, the continued monitoring of the antimicrobial resistance establishment of a surveillance system is urgently needed to prevent further dissemination in Iran.
Keywords: Klebsiella pneumoniae, Wound samples, Multiplex polymerase chain reaction, Iran
Copyright and License Information
© 2020 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Klebsiella pneumoniae is a gram-negative, aerobic, nonmotile bacilli and is a common cause of a wide range of infections in humans and animals. In addition, it is one of the most prevalent enteric bacteria responsible for up to 10% of all nosocomial infections and is also involved in pneumonia and urinary tract infections causing severe morbidity and mortality (1). A recent report is also available regarding the highly invasive K. pneumoniae that causes primary liver abscesses in humans (2). These invasive, abscess forming strains of K. pneumoniae are associated with the so-called hypermucoviscosity (HMV) phenotype, which is a bacterial colony trait identified by a positive string test. The HMV phenotype is found in K. pneumoniae expressing either the capsular serotypes K1 or K2. The K1 serotypes of K. pneumoniae have 2 potentially important genes of rmpA and magA. The first one is a transcriptional activator of colanic acid biosynthesis and the second one encodes a 43-kD outer membrane protein (3). Further, the serotype capsules of K1 and K2 can cause intense diseases and based on the studies on these serotypes, magA and rmpA genes, related to HMV “in charge of the positive synthesis of outside-capsule polysaccharide” are both useful tools in knowing such serotypes (4,5). Most K. pneumoniae isolates have a chromosomally encoded SHV-1 β-lactamase (6). Since 1983, plasmid-encoded extended-spectrum β-lactamases (ESBLs) derived from the TEM and SHV families have been extensively reported in some Gram-positive bacilli such as Staphylococcus aureus and Enterobacteriaceae, especially in Klebsiella spp. (7). Furthermore, the emergence and spread of multidrug-resistant K. pneumoniae, specifically the ESBL-producing strains, are often responsible for the failure of antibiotic treatment in hospital settings. However, the presence of resistance to trimethoprim-sulfamethoxazole can lead to treatment failure in cases of urinary tract infections in many countries (8). Sulfonamide resistance in Gram-negative bacilli generally arises from the acquisition of dihydropteroate synthase (DHPS) genes in integrons that are not inhibited by the drug. Three different types of DHPS genes have been currently identified, including sul1, sul2, and sul3 (9). The sul1 gene is found linked to other resistance genes in class 1 integrons and on large conjugative plasmids while sul2 is usually located on small nonconjugative plasmids, large transmissible multi-resistance plasmids, or through insertion element common region (ISCR2) element. sul3, as a plasmid-borne sulfonamide resistance gene, is also present although it is rare (10,11). Recent studies have shown that mobile and mobilizable DNA elements such as integrons play an important role in the development and dissemination of antibiotic resistance. Integrons are defined as site-specific recombination systems that are capable of integrating and expressing open reading frames contained in modular structures called mobile gene cassettes. Moreover, different classes of integrons are characterized by sequence differences in the intI gene encoding an integrase (12). Additionally, class 1 integrons possess two conserved segments (CSs), namely, the 5’-CS and the 3’-CS, which are separated by a variable region including the integrated antibiotic resistance gene cassettes of different lengths, arrangements, and sequences (13,14). Three main groups or classes of integrons associated with antibiotic resistance have been described in the clinical environment. Therefore, the present study investigated the genotypic and phenotypic antibiotic resistance patterns K. pneumoniae strains isolated from clinical samples in Iran.
Materials and Methods
Bacterial Strains and Identification
This study set out to determine 120 K. pneumoniae isolates which were detected from infected wound samples in Isfahan hospitals. All clinical isolates in addition to molecular serotyping were biochemically analyzed by conventional bacteriology tests.Furthermore,the polymerase chain reaction (PCR) method was used to detect the 16S-23S internal transcribed spacer unit of K. pneumoniae subsp. pneumoniae, facilitating the identification of the following subspecies:
F: ATTTGAAGAGGTTGCAAACGAT and R: TTCACT CTGAAGTTTTCTTGTTTC (amplicon size: 130 bp).
Further, cycling conditions were as follows:
Initial denaturation at 94°C for 5 minutes, 35 cycles of 94, 58, and 72°C each for 1 minute, respectively, followed by a final extension at 72°C for 7 minutes. K. pneumoniae ATCC13883 was used as the positive control as well (15).
Antimicrobial Susceptibility Testing
The antibiotic susceptibility was determined by the disk diffusion method on Mueller-Hinton agar plates (Merck, Darmstadt, Germany) as recommended by the Clinical Laboratory Standards Institute (CLSI). The disks containing the following antibiotics were used (Padtan-Teb, Iran):
Amoxicillin (10 μg), amikacin (30 μg), kanamycin (30 μg), tetracycline (30 μg), nalidixic acid (30 μg), co-trimoxazole (25 μg), ciprofloxacin (5 μg), cephalothin (30 μg), norfloxacin (10 μg), ceftriaxone (30 μg), nitrofurantoin (10 μg), imipenem (10 μg), cefepime (30 μg), and gentamicin (10 μg). Finally, Escherichia coli ATCC 25922 was used as quality control for the antimicrobial susceptibility test (15,16).
Polymerase Chain Reaction Assay
DNA template was extracted using the phenol and chloroform method and total DNA was measured at 260 nm optical density according to the method described by Sambrook and Russell. The reverse and forward primers, the size of the product for PCR programs, and volume, as previously published, were used for the detection of K. pneumoniae (17-19). The 1.5% agarose gel in the Tris-Borate-EDTA buffer was used for PCR product separation. Then, the gels were run at a constant voltage of 100 V for 1 hour, stained in 2 μg/mL ethidium bromide for 10 minutes, and photographed under UV by gel documentation system (20). Eventually, the reverse and forward primers, the size of the product for K. pneumoniae capsular serotypes were used (20).
Results
Serotyping and Antimicrobial Susceptibility Patterns of Klebsiella pneumonia
During the study, 120 K. pneumonia were isolated from wound samples in Isfahan hospitals in Iran, followed by performing molecular serotyping. Then, the disk diffusion method was used according to the CLSI (2017) in order to determine antibiotic resistance by the phenotypic pattern.
Of the total 120 K. pneumonia wound samples, 63 and 57 isolates were collected from females and males, respectively. In this research, the highest resistance was related to ceftazidime (74.3%), ciprofloxacin (78.5%), and tetracycline (72%). Furthermore, there was widespread resistance of the isolates to the antibiotic which is shown in Figure 1 (P<0.05).Tables 1 and 2 present the PCR results for capsular genes and the frequency of antimicrobial-related gens in K. pneumoniae.
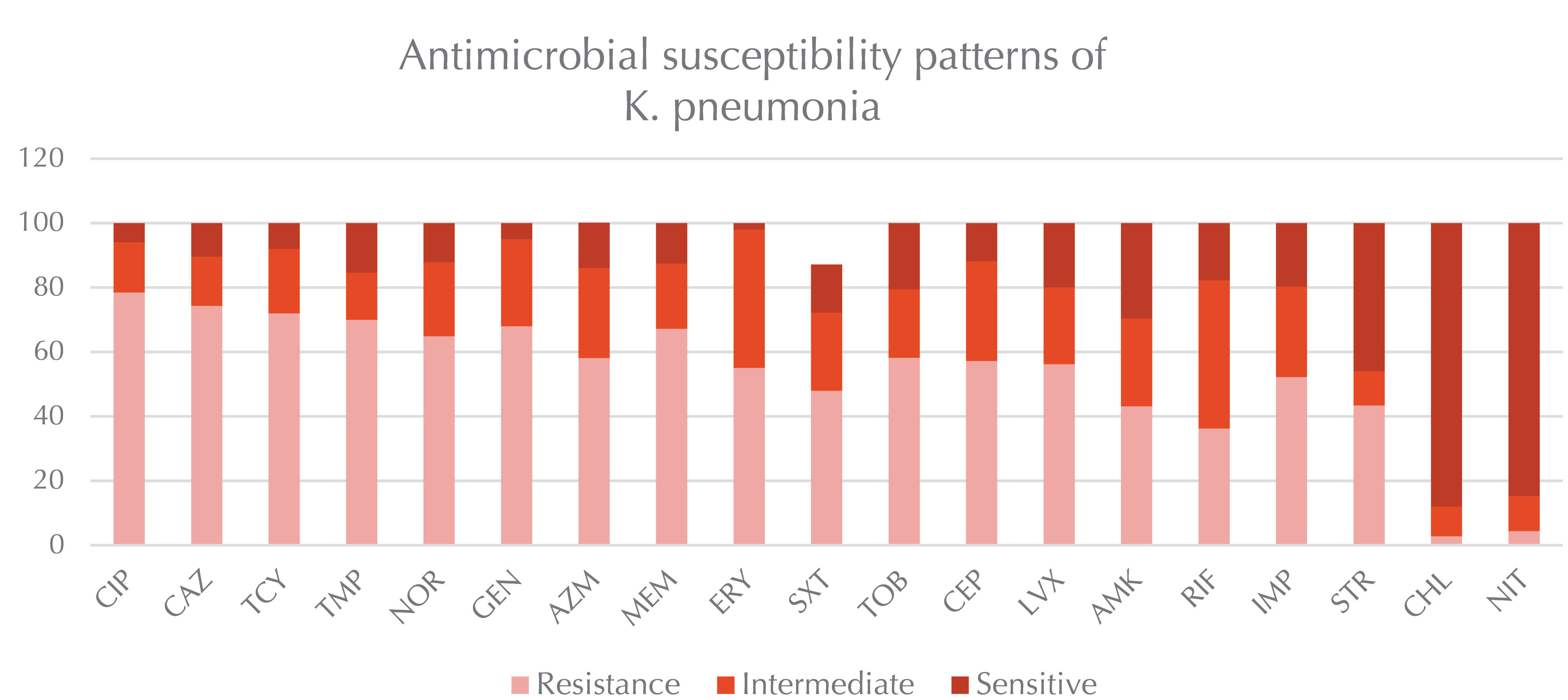
Figure 1.
Serotyping and Antimicrobial Susceptibility Patterns of Klebsiella pneumoniae.
.
Serotyping and Antimicrobial Susceptibility Patterns of Klebsiella pneumoniae.
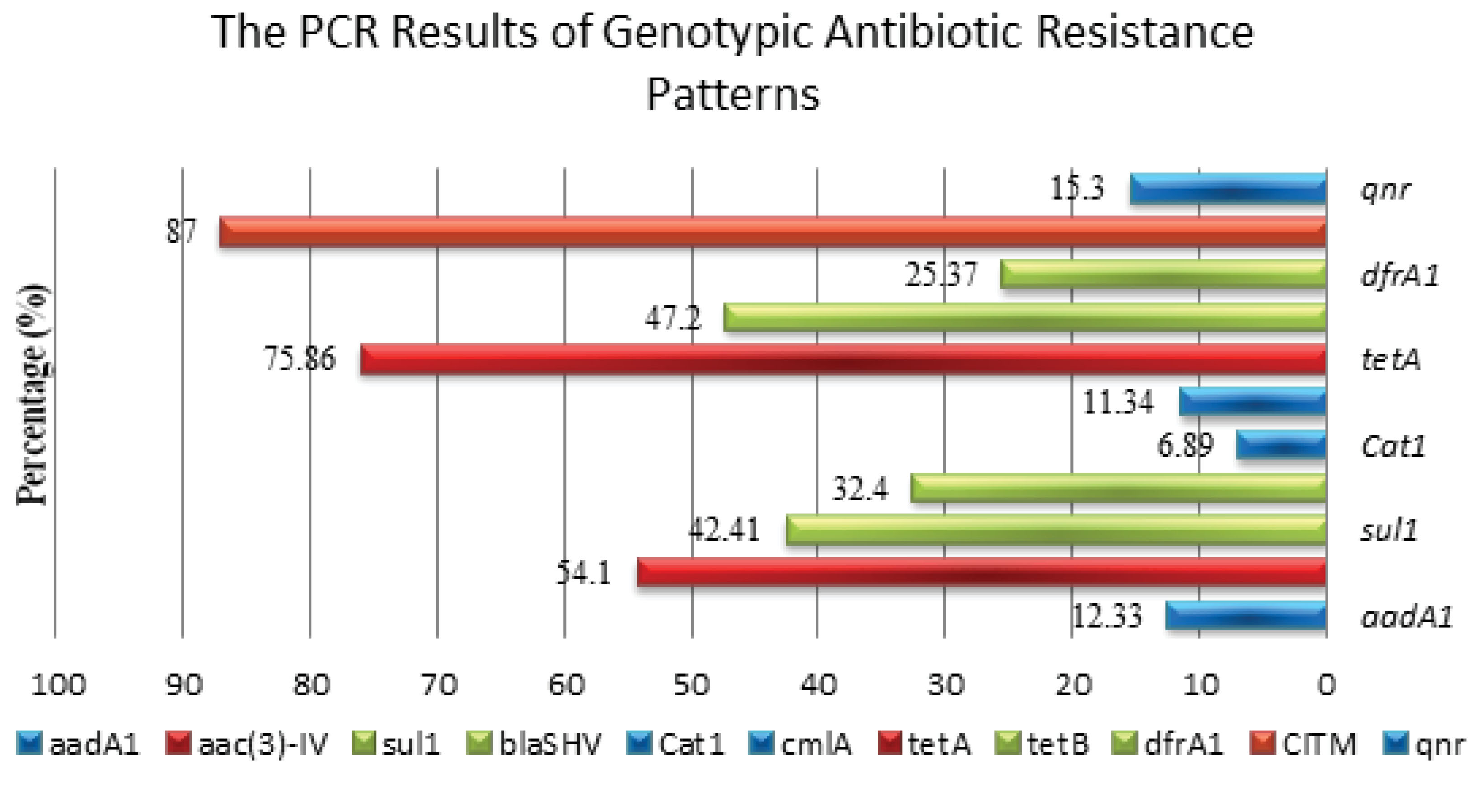
Figure 2.
Antimicrobial Susceptibility Patterns of Klebsiella pneumoniae.
.
Antimicrobial Susceptibility Patterns of Klebsiella pneumoniae.
Table 1.
The PCR Results (Frequency of Antimicrobial-Related Gens) for the Genotypic Antibiotic Resistance Patterns of Klebsiella pneumoniae
|
Gene
|
Size of
Product (bp)
|
Antimicrobial
Agent
|
Resistance Percentage by PCR (%)
|
|
aadA1
|
447 |
Streptomycin |
12.33 |
|
aac ( 3)-IV
|
286 |
Gentamicin |
54.1 |
|
sul1
|
822 |
Sulfonamides' |
42.41 |
|
blaSHV
|
768 |
Cephalothin |
62.4 |
|
Cat1
|
547 |
Chloramphenicol |
6.89 |
|
cmlA
|
698 |
Chloramphenicol |
11.34 |
|
tetA
|
577 |
Tetracycline |
75.86 |
|
tetB
|
634 |
Tetracycline |
47.2 |
|
dfrA1
|
367 |
Trimethoprim |
25.37 |
|
CITM
|
462 |
Ampicillin |
87 |
|
qnr
|
516 |
Fluoroquinolone |
15.3 |
Note. PCR: Polymerase chain reaction.
Table 2.
The PCR Results for Capsular Genes in Klebsiella pneumoniae
|
Gene for Capsule Type
|
Size of Product (bp)
|
Number of Samples
|
Percentage (%)
|
|
K1
|
1283 |
7 |
5.8 |
|
K2
|
641 |
78 |
78 |
|
K5
|
280 |
63 |
63 |
|
K54
|
881 |
23 |
19.16 |
|
K57
|
1037 |
13 |
10.83 |
Note. PCR: Polymerase chain reaction.
The levels of antibiotic resistance are shown in the samples. Based on the results, the highest resistance belonged to ceftazidime (74.3%), ciprofloxacin (78.5%), and tetracycline (72). The investigated antibiotics included ciprofloxacin, ceftazidime, tetracycline, trimethoprim, norfloxacin, gentamicin, azithromycin, meropenem, erythromycin, cotrimoxazole, tobramycin, cefalotin, levofloxacin, amikacin, rifampin, imipenem, streptomycin, chloramphenicol, and nitrofurantoin.
The PCR results represent the frequency of antimicrobial-related gens and the percentage for genotypic antibiotic resistance patterns in K. pneumoniae. According to the results, aac (3)-IV (63%) had the highest frequency among the examined genes.
Discussion
The results of the experiment found clear support for the multidrug-resistance patterns of K. pneumoniae, along with the frequency distribution of K. pneumoniae genes and their capsulargenes. Our findings with regard to the overall high resistance of K. pneumoniae strainsto antibiotics such as ceftazidime (74.3%), ciprofloxacin (78.5%), and tetracycline (72%) are in agreement with those of other recent studies. Similar to our results, Salimizand et al found the high rate of resistance to imipenem (100%) and meropenem (100%). Although other beta-lactam groups including aztreonam (20%), ceftazidime (20%), and cefotaxime (20%), indicated more antibiotic resistance, which contradicts our results (21). On the other hand, Mansury et al (22) reported that the highest rate of resistance belonged to amoxicillin (100%), cefotaxime (50%), and gentamicin (42.3%) and the lowest rates were observed for meropenem (11.8%), imipenem (15.9%), and amikacin (15.9%), which is in line with the results of previous studies.
The findings of the present study showed that K2 (78%) was the most common K. pneumoniae serotype, followed by K5 (63%). The obtained data further revealed that a total of 87% of K. pneumonia isolates carried CITM and 75.86%of themcarried tetA. This is contrary to the human isolates of K. pneumoniae in which the tetAgene is present in both K1 and K2 capsular serotypes, as well as nearly 67% of non-K1/K2 serotypes, but the cat1 gene appears restricted to the isolates of the K1 serotype. This result ties well with that of Jung-Chung, representing that poor glycemic control in diabetic patients plays a significant role in impairing the neutrophil phagocytosis of serotype K1/K2 K. pneumoniae (23). Khamesipour and Tajbakhsh (15) found a similar result and reported that serotype K1 is one of the most important serotypes of K. pneumonia and of 13 out of 90 isolates had serotype K1A (14.44%) and 15 cases had serotype K2A (16.66%).
Based on the findings of the current study, more than half of the K. pneumoniae strains possessed one or more of these sulgenes and sulfonamides and gentamicinresistance occurred in 42.41% and 54.1% of these strains, respectively. This result is in line with the findings of another study done among Escherichia coli strains where the sul2 gene was found to be predominant in E. coli strains isolated in urinary tract infection episodes. The results of this study confirm the need for increasing concern regarding therapy for clinical infections caused by K. pneumoniae isolates that have resistance-encoding CITM andsul and tetA genes which are related to class 1 integrons. Therefore, the continued monitoring of antimicrobial resistance, the adoption of the prudent use of antimicrobial agents and the establishment of a surveillance system is highly needed for preventing further dissemination in Iran (24).
Conclusions
In general, it is necessary to continuously monitor antimicrobial resistance, adopt the prudent use of antimicrobial agents, and establish a surveillance system to prevent further dissemination in Iran.
Conflict of Interests
The authors declared no conflict of interests.
Authors Contribution
FN: Ideas; evolution of overarching research goals and aims, preparation of the published work.
SB and DD: investigation process experiments, provision of study materials.
ET: Management and coordination responsibility for the research activity planning and execution, acquisition of the financial support for the project.
Funding
This study was financially supported by Islamic Azad University of Shahrekord, Iran, project number 97/3940.
References
- Vaez H, Sahebkar A, Khademi F. Carbapenem-resistant Klebsiella pneumoniae in Iran: a systematic review and meta-analysis. J Chemother 2019; 31(1):1-8. doi: 10.1080/1120009x.2018.1533266 [Crossref] [ Google Scholar]
- Tan TY, Ong M, Cheng Y, Ng LSY. Hypermucoviscosity, rmpA, and aerobactin are associated with community-acquired Klebsiella pneumoniae bacteremic isolates causing liver abscess in Singapore. J Microbiol Immunol Infect 2019; 52(1):30-4. doi: 10.1016/j.jmii.2017.07.003 [Crossref] [ Google Scholar]
- Ranjbar R, Fatahian Kelishadrokhi A, Chehelgerdi M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect Drug Resist 2019; 12:603-11. doi: 10.2147/idr.s199639 [Crossref] [ Google Scholar]
- Wang CH, Lu PL, Liu EY, Chen YY, Lin FM, Lin YT. Rapid identification of capsular serotype K1/K2 Klebsiella pneumoniae in pus samples from liver abscess patients and positive blood culture samples from bacteremia cases via an immunochromatographic strip assay. Gut Pathog 2019; 11:11. doi: 10.1186/s13099-019-0285-x [Crossref] [ Google Scholar]
- Ekwanzala MD, Dewar JB, Kamika I, Momba MNB. Tracking the environmental dissemination of carbapenem-resistant Klebsiella pneumoniae using whole genome sequencing. Sci Total Environ 2019; 691:80-92. doi: 10.1016/j.scitotenv.2019.06.533 [Crossref] [ Google Scholar]
- Pishtiwan AH, Khadija KM. Prevalence of blaTEM, blaSHV, and blaCTX-M genes among ESBL-producing Klebsiella pneumoniae and Escherichia coli Isolated from thalassemia patients in Erbil, Iraq. Mediterr J Hematol Infect Dis 2019; 11(1):e2019041. doi: 10.4084/mjhid.2019.041 [Crossref] [ Google Scholar]
- Nourbakhsh F, Momtaz H. Evaluation of phenotypic and genotypic biofilm formation in Staphylococcus aureus isolates isolated from hospital infections in Shahrekord, 2015. Journal of Arak University of Medical Sciences 2016; 19(4):69-79. [ Google Scholar]
- Smilack JD. Trimethoprim-sulfamethoxazole. Mayo Clin Proc 1999; 74(7):730-4. doi: 10.4065/74.7.730 [Crossref] [ Google Scholar]
- Grape M, Farra A, Kronvall G, Sundstrom L. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant gram-negative bacteria. Clin Microbiol Infect 2005; 11(3):185-92. doi: 10.1111/j.1469-0691.2004.01059.x [Crossref] [ Google Scholar]
- Perreten V, Boerlin P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother 2003; 47(3):1169-72. doi: 10.1128/aac.47.3.1169-1172.2003 [Crossref] [ Google Scholar]
- Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother 1995; 39(2):279-89. doi: 10.1128/aac.39.2.279 [Crossref] [ Google Scholar]
- Nourbakhsh F, Rajai M, Momtaz H. Antibiotic resistance and carriage integron classes in clinical isolates of Acinetobacter baumannii from Isfahan hospitals, Iran. Zahedan J Res Med Sci 2017; 19(1):e6009. doi: 10.17795/zjrms-6009 [Crossref] [ Google Scholar]
- Nourbakhsh F, Nourbakhsh V, Jafakesh MT. Prevalence of class I, II and III integrons in the antibiotic-resistant isolates of A baumannii detected from patients hospitalized in medical centers of Shahrekord. Feyz Journal of Kashan University of Medical Sciences 2016; 20(5):461-8. [ Google Scholar]
- Nourbakhsh F, Ebrahimzadeh Namvar A, Momtaz H. Characterization of staphylococcal cassette chromosome Mec elements in biofilm-producing Staphylococcus aureus, isolated from hospital infections in Isfahan. International Journal of Medical Laboratory 2016; 3(1):33-42. [ Google Scholar]
- Khamesipour F, Tajbakhsh E. Analyzed the genotypic and phenotypic antibiotic resistance patterns of Klebsiella pneumoniae isolated from clinical samples in Iran. Biomed Res 2016; 27(4):1017-26. [ Google Scholar]
- Tajbakhsh E, Tajbakhsh S, Khamesipour F. Isolation and molecular detection of gram negative bacteria causing urinary tract infection in patients referred to Shahrekord hospitals, Iran. Iran Red Crescent Med J 2015; 17(5):e24779. doi: 10.5812/ircmj.17(5)2015.24779 [Crossref] [ Google Scholar]
- Rudolph KM, Parkinson AJ, Black CM, Mayer LW. Evaluation of polymerase chain reaction for diagnosis of pneumococcal pneumonia. J Clin Microbiol 1993; 31(10):2661-6. [ Google Scholar]
- Campbell LA, Perez Melgosa M, Hamilton DJ, Kuo CC, Grayston JT. Detection of Chlamydia pneumoniae by polymerase chain reaction. J Clin Microbiol 1992; 30(2):434-9. [ Google Scholar]
- Manchanda V, Rai S, Gupta S, Rautela RS, Chopra R, Rawat DS. Development of TaqMan real-time polymerase chain reaction for the detection of the newly emerging form of carbapenem resistance gene in clinical isolates of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii. Indian J Med Microbiol 2011; 29(3):249-53. doi: 10.4103/0255-0857.83907 [Crossref] [ Google Scholar]
- Yu WL, Fung CP, Ko WC, Cheng KC, Lee CC, Chuang YC. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 2007; 195(8):1235-6. doi: 10.1086/512686 [Crossref] [ Google Scholar]
- Salimizand H, Shahcheraghi F, Kalantar E, Badmasti F, Mousavi SF. Molecular characterization of class 1 integrons and gene cassettes in multidrug resistant (MDR) Klebsiella spp isolated from hospitalized and outpatients in Iran, 2009. Iran J Microbiol 2013; 5(1):48-55. [ Google Scholar]
- Mansury D, Motamedifar M, Sarvari J, Shirazi B, Khaledi A. Antibiotic susceptibility pattern and identification of extended spectrum beta-lactamases (ESBLs) in clinical isolates of Klebsiella pneumoniae from Shiraz, Iran. Iran J Microbiol 2016; 8(1):55-61. [ Google Scholar]
- Lin JC, Siu LK, Fung CP, Tsou HH, Wang JJ, Chen CT. Impaired phagocytosis of capsular serotypes K1 or K2 Klebsiella pneumoniae in type 2 diabetes mellitus patients with poor glycemic control. J Clin Endocrinol Metab 2006; 91(8):3084-7. doi: 10.1210/jc.2005-2749 [Crossref] [ Google Scholar]
- Arabi H, Pakzad I, Nasrollahi A. Sulfonamide Resistance Genes (sul) M in Extended Spectrum Beta Lactamase (ESBL) and Non-ESBL Producing Escherichia coli Isolated From Iranian Hospitals. Jundishapur J Microbiol 2015; 8(7):e19961. doi: 10.5812/jjm.19961v2 [Crossref] [ Google Scholar]