Avicenna Journal of Clinical Microbiology and Infection. 6(3):83-87.
doi: 10.34172/ajcmi.2019.15
Original Article
Evaluation of the Prevalence of blaSHV, blaTEM, and blaCTX Genes in Escherichia coli Isolated From Urinary Tract Infections
Majid Alipour 1, *  , Ameneh Jafari 1
, Ameneh Jafari 1
Author information:
1Department of Cell and Molecular Biology, Babol Branch, Islamic Azad University, Babol, Iran
*
Corresponding author: Majid Alipour, Ph.D. in Cellular and Molecular Biology, Assistant professor, Department of Cell and Molecular Biology, Islamic Azad University of Babol Branch, Babol, Iran Email:
alipourmk@gmail.com
Abstract
Background: Beta-lactamases are the most important factors in the resistance to beta-lactam antibiotics among gram-negative bacteria, especially Escherichia coli. Today, the prevalence of infections caused by extended-spectrum β-lactamases (ESBLs)-producing E. coli is increasing, as one of the emerging health problems worldwide. This study aimed to investigate the prevalence of blaSHV (sulfhydryl variable β-lactamase), blaTEM (temoneira β-lactamase), and blaCTX (cefotaximase β-lactamase) genes in E. coli isolated from urinary tract infections (UTIs).
Methods: In this study, 3192 midstream urine samples collected from Babol and Qaemshahr counties, Mazandaran province (Iran) were cultured on eosin methylene blue and blood agars. An antibiotic susceptibility test was performed to determine ESBL-producing E. coli isolates using the combined disk method. Finally, the ESBLs were evaluated for the presence of
blaSHV, blaTEM, and blaCTX genes by the polymerase chain reaction (PCR) technique.
Results: Of the 3192 cultured urine samples, 192 isolates were identified as E. coli by the IMViC and biochemical tests. In addition, the ESBL producers were detected in 45 (28/12 %) out of 192 E. coli isolates by the double-blind synergism test. The PCR of the 45 ESBL-producing E. coli isolates demonstrated that the blaTEM was the most abundant gene (89%), followed by blaCTX-M (27%) and blaSHV (20%). Eventually, the co-existence of
blaSHV, blaCTX-M, and blaTEM was detected in 3 (7%) isolates.
Conclusions: Due to the high prevalence of ESBL-producing uropathogenic E. coli. (UPEC) in the studied region, future studies are recommended to perform phenotypic or genotypic tests to detect ESBL-producing isolates in laboratories to select appropriate antibiotics for treating UTIs.
Keywords: Extended-spectrum β-lactamases, Uropathogenic Escherichia coli, Urinary tract infection
Copyright and License Information
© 2019 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Escherichia coli is the most important agent of urinary tract infections (UTIs) which accounts for nearly 75% of the isolates (1,2). Annually, about 150 million women suffer from UTIs worldwide (3). Uropathogenic E. coli (UPEC) strains often produce α-hemolysin which can cause an inflammatory response by the lysis of cells and the release of cytokine (4). β-lactamases are the enzymes that disrupt beta-lactam antibiotics and secret into the periplasmic space in gram-negative bacteria or out of the cell by Gram-positive organisms (5,6). Based on the protein sequence homology (the Ambler classification scheme), beta-lactamases are divided into A, B, C, and D types. According to Tooke et al study (7), the A-, C-, and D-type β-lactamases have serine at their active site (seine-β-lactamases) whereas B-type enzymes are considered as zinc-containing metalloenzymes (metallo-β-lactamases). The extended-spectrum β-lactamases (ESBLs) were first recognized in Klebsiella pneumoniae and Serratia marcescens in the mid-1980s. These enzymes are produced by gram-negative bacilli and are often classified into temoneira (blaTEM β-lactamase), sulfhydryl variable active site (blaSHV β-lactamase), and the cefotaxime degrading enzyme (blaCTX-M β-lactamase) classes (8). The high prevalence of ESBLs in E. coli has raised concerns about the treatment of the infection caused by this bacterium (9). The ESBLs represent resistance to almost all β-lactam antibiotics except for carbapenems and cefamycins and are mainly produced by E. coli and K. pneumoniae (10). TEM-1, TEM-2, and SHV-1 are named narrow-spectrum β-lactamases because they hydrolyze penicillins and are considered as the first generation of cephalosporins such as cephalothin, cephaloridine, or cefazolin (11). The ESBLs are emanated from the narrow-spectrum beta-lactamase genes (i.e., TEM-1, TEM-2, or SHV-1) by mutations that change the amino acid sequence at their active site (12). The genes encoding ESBLs are located on the bacterial chromosome or plasmid (13). The blaSHV, blaTEM, and blaCTX-M types are regarded as the most common ESBLs (14). The UPEC strains contain blaSHV and blaTEM genes which cause resistance to beta-lactam antibiotics and the bacterium also possesses a blaCTX-M gene that hydrolyzes cefotaxime and ceftazidime (15). Given the importance of resistance to penicillins, cephalosporins, and carbapenems, mediated by extended-spectrum β-lactamases and carbapenemases, this study was performed to investigate the prevalence of ESBL enzymes in the UPEC strainsisolated from patients with UTIs.
Materials and Methods
Isolation and Detection of Escherichia coli
This study was conducted on 3192 patients who referred to the therapeutic centers of Babol and Qaemshahr, Mazandaran Province, Iran between March and December 2017. The urine samples were streaked on eosin methylene blue and blood agars (Merk Company, Germany) and placed at 37°C for 24 hours. UPEC strains were identified using the gram-stain, the IMViC test, b-hemolytic activity, and the other conventional biochemical tests. These strains were then stored in Luria-Bertani (LB) broth (Merk Company, Germany) including 20% glycerol at -20°C for future studies (16).
Combined Disc Test for the Phenotypic Detection of Extended-Spectrum β -Lactamases
One hundred and ninety-two E. coli isolates were studied to detect the presence of ESBLs by the combined disc method. A 0.5 McFarlandsuspension of E. coli was spread on the Mueller-Hinton agar. In this study, ceftazidime (30 μg) disks (MAST, UK) alone and in the combination of ceftazidime and clavulanic acid (30/10 μg) disks (MAST, UK) were placed on the Mueller-Hinton agar at a distance of 20 mm from each other and were placed at 3 °C for 24 hours. The isolates that displayed an increase of ≥5 mm in the inhibition halo of the combined disk (ceftazidime plus clavulanic acid) were considered as an ESBL producer compared to the ceftazidim disc alone (17,18). The polymerase chain reaction (PCR) detection was carried out on all positive ESBLs. The isolates were confirmed by the PCR and the Basic Local Alignment Search Tool (BLAST) sequence analysis was used as a positive control.
Extraction of DNA by Boiling Lysis Method
A single colony was used for inoculating 5 mL of LB broth and then was incubated at 37°C for 24 hours. Next, 1 mL of bacterial suspension was transferred to a 1.5 mL microcentrifuge tube and was centrifuged at 6000 rounds per minute (rpm) for 5 minutes. The supernatant fluid was discarded as well. The pellet was resuspended in 200 μL nuclease-free distilled water and boiled for 10 minutes and chilled immediately on the ice for 5 minutes. After the ice incubation, the tubes were centrifuged at 10 000 rpm for 5 minutes at 4°C and the supernatant was transferred into a new tube. An aliquot of 3 μL of the supernatant was used in the PCR mixes (19). This method of DNA purification was selected to harvest both plasmid and chromosomal DNA.
Genotypic Detection of ESBL Genes
All the ESBL-producing E. coli strains confirmed by the phenotypic assay were screened using the uniplex PCR for the detection of blaTEM, blaSHV, and blaCTX-M genes. The PCRs were carried out in a final volume of 25 μL. The oligonucleotide primers used in the investigation are listed in Table 1.
Table 1.
The Primers Used in This Investigation
|
Gene
|
Primer Sequence
5’→3’
|
Amplicon Size (bp)
|
Annealing Temperature
|
Reference
|
TEM -F
TEM-R |
TTGGGTGCACGAGTGGGTTA
TAATTGTTGCCGGGAAGCTA |
500 |
52 |
(20) |
SHV-F
SHV-R |
AGGATTGACTGCCTTTTTG
ATTTGCTGATTTCGCTCG |
392 |
54 |
(21) |
CTX-M-F
CTX-M-R |
ACCGCCGATAATTCGCAGAT
GATATCGTTGGTGGTGCCATAA |
585 |
58 |
(22) |
The PCRs were performed with a 25 𝜇L reaction mixture containing 3 𝜇L of the solution containing DNA, 12.5 𝜇L of super PCR master mix 2X (Yekta Tajhiz Azma), 1 𝜇L of each primer (20 pmol), and 7.5 𝜇L of distilled water. In addition, the PCR was perfor.for 5 minutes. The BLAST analysis revealed that the PCR product sequences all 3 genes show high similarity with the corresponding genes in the GenBank database thus confirming the genes. A clinical isolate containingblaTEM, blaSHV, and blaCTX genes confirmed by the PCR and sequencing techniques were used as positive control and sterile distilled water was utilized as a negative control. The PCR products were identified on 2% agarose gel electrophoresis and the gels were stained with ethidium bromide and then were visualized by the UV trans-illuminator (20,21,22).
Data Analysis
The data were analyzed by SPSS software, version 16. The prevalence of resistance genes was calculated using c2and fisher extract tests for each gene. Statistical significance was considered at the P< 0.05.
Results
The urine samples were collected from hospitalized patients (inpatients) and outpatients. Of the 3192 urine samples cultured on the eosin methylene blue and blood agars, 192 isolates were identified as E. coli by IMViC and biochemical tests. Among these 192 E. coli strains, the ESBL producers were detected in 45 cases (28/12%) by the combined disc test. Figures 1, 2, and 3 illustrate the PCR product bands of blaTEM, blaSHV, and blaCTX-M genes.
The amplified PCR products forblaTEM, blaSHV, andblaCTX-M genes were 500, 392, and 585 base pairs (bp), respectively. All the 45 E. coli isolates confirmed by the phenotypic methods were also positive for at least one ESBL gene by the molecular technique (P < 0.001), the details of which are provided in Table 2.
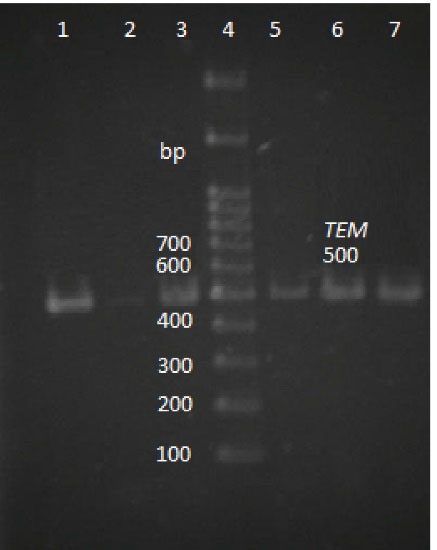
Figure 1.
Agarose Gel Electrophoresis of the PCR Products of blaTEM Gene (500 bp).
Note. Lane 4: DNA ladder; Lane 1: Positive control; Lane 2: Negative control; Lanes 3, 5, 6, 7, and 8: positive samples; PCR: Polymerase chain reaction.
.
Agarose Gel Electrophoresis of the PCR Products of blaTEM Gene (500 bp).
Note. Lane 4: DNA ladder; Lane 1: Positive control; Lane 2: Negative control; Lanes 3, 5, 6, 7, and 8: positive samples; PCR: Polymerase chain reaction.
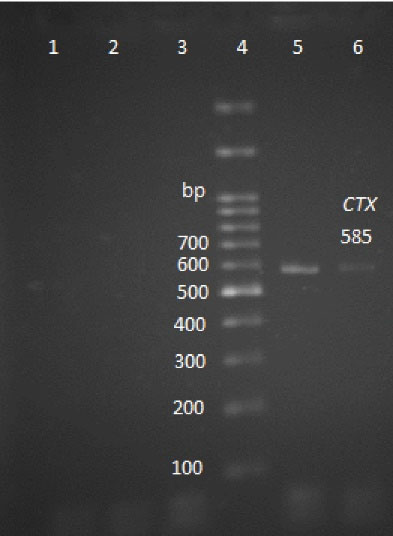
Figure 2.
Agarose Gel Electrophoresis of the PCR Products of blaSHV Gene (392 bp).
Note. Lanes 1, 2, 3, and 7: Positive samples; Lane 4: DNA ladder; Lane 6: Negative control; Lane 5: Positive control; PCR: Polymerase chain reaction.
.
Agarose Gel Electrophoresis of the PCR Products of blaSHV Gene (392 bp).
Note. Lanes 1, 2, 3, and 7: Positive samples; Lane 4: DNA ladder; Lane 6: Negative control; Lane 5: Positive control; PCR: Polymerase chain reaction.
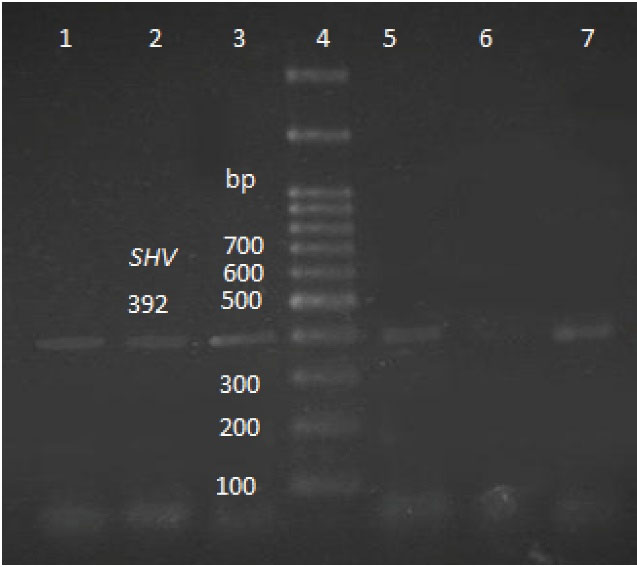
Figure 3.
Agarose Gel Electrophoresis of the PCR Products of blaCTX-M Gene (585 bp).
Note. Lanes 1 and 2: Negative samples; Lane 3: Negative control; Lane 4: DNA ladder; Lane 5: Positive control; Lane 6: Positive sample; PCR: Polymerase chain reaction.
.
Agarose Gel Electrophoresis of the PCR Products of blaCTX-M Gene (585 bp).
Note. Lanes 1 and 2: Negative samples; Lane 3: Negative control; Lane 4: DNA ladder; Lane 5: Positive control; Lane 6: Positive sample; PCR: Polymerase chain reaction.
Table 2.
Prevalence of ESBL Genes Among 54 Isolates
|
ESBL Genes
|
Number
|
Percent
|
|
bla
TEM
|
40 |
89 |
|
bla
CTX-M
|
12 |
27 |
|
bla
SHV
|
9 |
20 |
|
bla
CTX-M andblaTEM |
9 |
20 |
|
bla
SHV
and bla
TEM
|
5 |
12 |
|
bla
SHV
,bla
CTX-MandblaTEM |
3 |
7 |
Note. ESBL: Extended-spectrum β-lactamases.
The uniplex PCR of the 45 ESBL-producing E. coli strains indicated that the blaTEM was the most abundant gene (89%), followed by CTX-M (27%) and SHV (20%). The co-existence of the blaCTX-M andblaTEM, as well as blaSHV and blaTEMgenes was detected in 9 (20%) and 5 (12%) isolates, respectively. The co-existence of all 3 genes (i.e., blaSHV, blaCTX-M,andblaTEM) were observed in 3 isolates (7%).
Discussion
Cephalexin or amoxicillin/clavulanate is considered as one of the first options in the treatment of acute uncomplicated cystitis and can be prescribed in the treatment of cystitis caused by gram-positive cocci because it is effective against enterococci and staphylococci (23). The ESBL-producing bacteria, particularly E. coli and K. pneumoniae has emerged as a significant problem in the treatment of bacterial infections worldwide (24). The value of the current study was 0.001 thus the results demonstrated that there is a significant relationship between phenotypic and genotypic methods for the detection of the ESBLs. In our study, the prevalence of ESBL-producing E. coli strains was 28.12%. Further, Yılmaz et al. found that the prevalence of ESBL-producing E. coli strains was 24% (25). In another study, Jena et al reported that the prevalence of ESBL-producing E. coli strains isolated from patients with UTIs was 59.74% (26). Although mutations can cause antibiotic-resistant, the overuse of antibiotics increases the selection and emergence of resistant bacteria strains. Therefore, the prevalence of ESBL-producing UPEC varies in different regions. In the present study, 40 (89%) isolates were positive for the blaTEM gene, which is consistent with the results of several studies. For example, Liu et al reported a prevalence of 72.1% for the blaTEM genotype (27). Furthermore, Jena et al (26) showed that among ESBL-producing E. coli isolated from patients with UTI, the prevalence of blaTEM gene was the predominant (93.47%). In the current study, the detection rate for the blaCTX-M gene was 12 (27%). In studies conducted by Ruppé et al and Majeed et al, the detection rate of the blaCTX-M gene in patients with UTI was 26 (76.4 %) and 26%, respectively (28,29). The result of our research is nearly similar to the report of Majeed et al, but it differs from that of Ruppé et al. These contradictory results indicate that the prevalence of the blaCTX-M gene varies in different regions. Moreover, the results of our study revealed that the prevalence of blaSHV was 9 (20%). According to Seyedjavadi et al and Reid et al, the rate of isolation of the blaSHV gene in UPEC was 45% and 2.4%, respectively (30,31). The reason for the different rates of the blaSHV is that the prevalence varies in various areas. Additionally, our finding showed that 39% of the UPEC isolates contained 2 or more ESBL genes. Similarly, Seyedjavadi et al (30) and Manoharan et al (32) reported the co-existence of different ESBL genes within the same isolate. Based on previous evidence, the co-existence of ESBL genes (i.e., blaSHV, blaCTX-M, and blaTEM) is because they are frequently located on the plasmid and can be transferred to other bacteria (33).
Conclusions
Due to the high prevalence of the ESBL uropathogenic E. coli, especially blaTEM (89%), it is thought that ESBLs should be detected by phenotypic or genotypic methods for selecting the appropriate antibiotics regarding treating the patients with UTIs. Thus, future studies are suggested to determine the prevalence of the metallo-b-lactamases of the Verona Integron-encoded metallo-β-lactamase, imipenemases, and New Delhi metallo-β-lactamase (16,34).
Ethical Approval
There are no ethical issues for this article.
Conflict of Interests
Authors declare no conflict of interests associated with this study.
Acknowledgments
The authors would like to thank Zohreh Eslamdost from Yahyanejad hospital for her excellent technical assistance.
References
- Smart A, de Lacy Costello B, White P, Avison M, Batty C, Turner C. Sniffing out resistance - Rapid identification of urinary tract infection-causing bacteria and their antibiotic susceptibility using volatile metabolite profiles. J Pharm Biomed Anal 2019; 167:59-65. doi: 10.1016/j.jpba.2019.01.044 [Crossref] [ Google Scholar]
- Lakshminarayana SA, Chavan SKD, Prakash R, Sangeetha S. Bacterial pathogens in urinary tract infection and antibiotic susceptibility pattern from a Teaching Hospital, Bengaluru, India. Int J Curr Microbiol Appl Sci 2015; 4(11):731-6. [ Google Scholar]
- De Nisco NJ, Neugent M, Mull J, Chen L, Kuprasertkul A, de Souza Santos M. Direct Detection of tissue-resident bacteria and chronic inflammation in the bladder wall of postmenopausal women with recurrent urinary tract infection. J Mol Biol 2019; 431(21):4368-79. doi: 10.1016/j.jmb.2019.04.008 [Crossref] [ Google Scholar]
- Söderström CM, Fagerberg SK, Brogaard MB, Leipziger J, Skals M, Praetorius HA. Loop diuretics diminish hemolysis induced by alpha-hemolysin from Escherichia coli. J Membr Biol 2017; 250(3):301-13. doi: 10.1007/s00232-017-9963-0 [Crossref] [ Google Scholar]
- Bellio P, Luzi C, Mancini A, Cracchiolo S, Passacantando M, Di Pietro L. Cerium oxide nanoparticles as potential antibiotic adjuvant Effects of CeO2 nanoparticles on bacterial outer membrane permeability. Biochim Biophys Acta Biomembr 2018; 1860(11):2428-35. doi: 10.1016/j.bbamem.2018.07.002 [Crossref] [ Google Scholar]
- Samaha-Kfoury JN, Araj GF. Recent developments in beta lactamases and extended spectrum beta lactamases. BMJ 2003; 327(7425):1209-13. doi: 10.1136/bmj.327.7425.1209 [Crossref] [ Google Scholar]
- Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y. beta-Lactamases and beta-lactamase inhibitors in the 21st century. J Mol Biol 2019; 431(18):3472-500. doi: 10.1016/j.jmb.2019.04.002 [Crossref] [ Google Scholar]
- Al-Jamei SA, Albsoul AY, Bakri FG, Al-Bakri AG. Extended-spectrum beta-lactamase producing E coli in urinary tract infections: A two-center, cross-sectional study of prevalence, genotypes and risk factors in Amman, Jordan. J Infect Public Health 2019; 12(1):21-5. doi: 10.1016/j.jiph.2018.07.011 [Crossref] [ Google Scholar]
- Mazzariol A, Bazaj A, Cornaglia G. Multi-drug-resistant gram-negative bacteria causing urinary tract infections: a review. J Chemother 2017; 29(sup1):2-9. doi: 10.1080/1120009x.2017.1380395 [Crossref] [ Google Scholar]
- Namikawa H, Yamada K, Yamairi K, Shibata W, Fujimoto H, Takizawa E. Mortality caused by extended-spectrum beta-lactamase-producing Enterobacteriaceae bacteremia; a case control study: alert to Enterobacteriaceae strains with high minimum inhibitory concentrations of piperacillin/tazobactam. Diagn Microbiol Infect Dis 2019; 94(3):287-92. doi: 10.1016/j.diagmicrobio.2019.01.018 [Crossref] [ Google Scholar]
- Lee J, Oh CE, Choi EH, Lee HJ. The impact of the increased use of piperacillin/tazobactam on the selection of antibiotic resistance among invasive Escherichia coli and Klebsiella pneumoniae isolates. Int J Infect Dis 2013; 17(8):e638-43. doi: 10.1016/j.ijid.2013.01.030 [Crossref] [ Google Scholar]
- Hassan MM, Gaber A, Alsanie WF, El-Hallous EI, Mohamed AA, Alharthi AA. Phylogeny and detection of blaTEM, blaSHV, blaCTX-M genes in Escherichia coli isolates from patients with urinary tract infections in Taif hospitals, Saudi Arabia. Ann Res Rev Biol 2018; 25(1):1-8. doi: 10.9734/ARRB/2018/39689 [Crossref] [ Google Scholar]
- Mondal AH, Siddiqui MT, Sultan I, Haq QMR. Prevalence and diversity of blaTEM, blaSHV and blaCTX-M variants among multidrug resistant Klebsiella spp from an urban riverine environment in India. Int J Environ Health Res 2019; 29(2):117-29. doi: 10.1080/09603123.2018.1515425 [Crossref] [ Google Scholar]
- Guzmán M, Salazar E, Cordero V, Castro A, Villanueva A, Rodulfo H. Multidrug resistance and risk factors associated with community-acquired urinary tract infections caused by Escherichia coli in Venezuela. Biomedica 2019; 39(s1):96-107. doi: 10.7705/biomedica.v39i2.4030 [Crossref] [ Google Scholar]
- Bravata-Alcantara JC, Bello-Lopez JM, Cortes-Ortiz IA, Mendez-Velazquez JJ, Aviles-Soto B, Quintas-Granados LI. Distribution of virulence and antimicrobial resistance genes in phylogenetic groups of Escherichia coli strains isolated from Mexican patients with urinary infection. Jundishapur J Microbiol 2019; 12(3):e83711. doi: 10.5812/jjm.83711 [Crossref] [ Google Scholar]
- Shams S, Hashemi A, Esmkhani M, Kermani S, Shams E, Piccirillo A. Imipenem resistance in clinical Escherichia coli from Qom, Iran. BMC Res Notes 2018; 11(1):314. doi: 10.1186/s13104-018-3406-6 [Crossref] [ Google Scholar]
- Parajuli NP, Maharjan P, Parajuli H, Joshi G, Paudel D, Sayami S. High rates of multidrug resistance among uropathogenic Escherichia coli in children and analyses of ESBL producers from Nepal. Antimicrob Resist Infect Control 2017; 6:9. doi: 10.1186/s13756-016-0168-6 [Crossref] [ Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: 28th informational supplement. Wayne, PA: CLSI; 2018.
- Das S, Dash HR. Microbial Biotechnology- A Laboratory Manual for Bacterial Systems. India: Springer; 2014.
- Tzelepi E, Magana C, Platsouka E, Sofianou D, Paniara O, Legakis NJ. Extended-spectrum beta-lactamase types in Klebsiella pneumoniae and Escherichia coli in two Greek hospitals. Int J Antimicrob Agents 2003; 21(3):285-8. doi: 10.1016/s0924-8579(02)00361-8 [Crossref] [ Google Scholar]
- Bhattacharjee A, Sen MR, Prakash P, Anupurba S. Role of beta-lactamase inhibitors in enterobacterial isolates producing extended-spectrum beta-lactamases. J Antimicrob Chemother 2008; 61(2):309-14. doi: 10.1093/jac/dkm494 [Crossref] [ Google Scholar]
- Ezeanya C, Agbakoba N, Ejike C, Okwelogu S. Evaluation of a Chromogenic Medium for the Detection of ESBL with Comparison to Double Disk Synergy Test. Br J Med Med Res 2017; 21(12):1-11. doi: 10.9734/BJMMR/2017/33259 [Crossref] [ Google Scholar]
- Choe HS, Lee SJ, Yang SS, Hamasuna R, Yamamoto S, Cho YH. Summary of the UAA-AAUS guidelines for urinary tract infections. Int J Urol 2018; 25(3):175-85. doi: 10.1111/iju.13493 [Crossref] [ Google Scholar]
- Chong Y, Shimoda S, Shimono N. Current epidemiology, genetic evolution and clinical impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae. Infect Genet Evol 2018; 61:185-8. doi: 10.1016/j.meegid.2018.04.005 [Crossref] [ Google Scholar]
- Yılmaz N, Ağuş N, Bayram A, Şamlıoğlu P, Şirin MC, Derici YK. Antimicrobial susceptibilities of Escherichia coli isolates as agents of community-acquired urinary tract infection (2008-2014). Turk J Urol 2016; 42(1):32-6. doi: 10.5152/tud.2016.90836 [Crossref] [ Google Scholar]
- Jena J, Sahoo RK, Debata NK, Subudhi E. Prevalence of TEM, SHV, and CTX-M genes of extended-spectrum beta-lactamase-producing Escherichia coli strains isolated from urinary tract infections in adults. 3 Biotech 2017; 7(4):244. doi: 10.1007/s13205-017-0879-2 [Crossref] [ Google Scholar]
- Liu H, Wang Y, Wang G, Xing Q, Shao L, Dong X. The prevalence of Escherichia coli strains with extended spectrum beta-lactamases isolated in China. Front Microbiol 2015; 6:335. doi: 10.3389/fmicb.2015.00335 [Crossref] [ Google Scholar]
- Ruppé E, Hem S, Lath S, Gautier V, Ariey F, Sarthou JL. CTX-M beta-lactamases in Escherichia coli from community-acquired urinary tract infections, Cambodia. Emerg Infect Dis 2009; 15(5):741-8. doi: 10.3201/eid1505.071299 [Crossref] [ Google Scholar]
- Majeed HT, Aljanaby AAJ. Antibiotic Susceptibility Patterns and Prevalence of Some Extended Spectrum Beta-Lactamases Genes in Gram-Negative Bacteria Isolated from Patients Infected with Urinary Tract Infections in Al-Najaf City, Iraq. Avicenna J Med Biotechnol 2019; 11(2):192-201. [ Google Scholar]
- Seyedjavadi SS, Goudarzi M, Sabzehali F. Relation between blaTEM, blaSHV and blaCTX-M genes and acute urinary tract infections. J Acute Dis 2016; 5(1):71-6. [ Google Scholar]
- Reid R, Al-Bayati M, Samarasinghe S. Genotypic Identification of Extended-Spectrum beta-Lactamase (ESBL)Producing Enterobacteriaceae from Urinary Tract Infections in the Leicestershire Area, United Kingdom: A One Health Prospective. J Infect Dis Diagn 2018; 3(2):122. doi: 10.4172/2576-389X.1000122 [Crossref] [ Google Scholar]
- Manoharan A, Premalatha K, Chatterjee S, Mathai D. Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol 2011; 29(2):161-4. doi: 10.4103/0255-0857.81799 [Crossref] [ Google Scholar]
- Gautam V, Thakur A, Sharma M, Singh A, Bansal S, Sharma A. Molecular characterization of extended-spectrum beta-lactamases among clinical isolates of Escherichia coli & Klebsiella pneumoniae: a multi-centric study from tertiary care hospitals in India. Indian J Med Res 2019; 149(2):208-15. doi: 10.4103/ijmr.IJMR_172_18 [Crossref] [ Google Scholar]
- Yoon EJ, Kang DY, Yang JW, Kim D, Lee H, Lee KJ. New Delhi metallo-beta-lactamase-producing Enterobacteriaceae in South Korea between 2010 and 2015. Front Microbiol 2018; 9:571. doi: 10.3389/fmicb.2018.00571 [Crossref] [ Google Scholar]