Avicenna Journal of Clinical Microbiology and Infection. 6(2):61-65.
doi: 10.34172/ajcmi.2019.12
Original Article
Prevalence of Coxiella burnetii in Traditional and Industrial Butter and Cream Using Nested Polymerase Chain Reaction in Shahrekord, Iran
Maryam Reisi 1, *, Ebrahim Rahimi 2, Manouchehr Momeni 1
Author information:
1Young Researchers and Elite Club, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran
2Department of Food Hygiene, College of Veterinary Medicine, Shahrekord Branch, Islamic Azad University,Shahrekord, Iran
*
Corresponding author: Maryam Reisi, Young Researchers and Elite Club, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran, Islamic Azad University, Rahmatieh, Shahrekord, Iran. Tel: 09139813707, Email:
maryam_r335@yahoo.com
Abstract
Background: A zoonotic disease of global importance known as "Q fever" is caused by Coxiella burnetii agent. The aim of the present research was to determine C. burnetii in the traditional and industrial butter and cream samples in Shahrekord, Iran.
Methods: This cross-sectional descriptive study was conducted from March to September 2016. Totally, 200 traditional and industrial butter and cream samples were collected from retailers in different regions of Shahrekord and then tested for C. burnetii via nested polymerase chain reaction (PCR) assays.
Results: In this survey, a total of 6 out of 200 samples (3%) were found to be positive for C. burnetii. More precisely, 4 (5.79%), 1 (5%), and 1 (2.56%) samples were related to the traditional bovine cream, traditional sheep butter, and traditional bovine butter samples, respectively. Nevertheless, no C. burnetii infections were found in the industrial butter and cream samples.
Conclusions: These results proved that traditional dairy products can be considered as an important reservoir for C. burnetii infection in Iran.
Keywords: Coxiella burnetii, Industrial butter, Industrial cream, Nested PCR, Iran
Copyright and License Information
© 2019 The Author(s); Published by Hamadan University of Medical Sciences.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (
http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium provided the original work is properly cited.
Background
Coxiella burnetiiis an agent of zoonosis diseases that occur in humans and animals. “Q fever” is the disease that is caused by C. burnetiiand coxiellosis, and affects humans and animals (1). C. burnetiias a small gram-negative bacteriumis morphologically similar to Rickettsia although they are genetically and physiologically different (2).Genus Coxiella has different species while C. burnetii is the only species of C. burnetiiformed via 16s-rRNA gene sequencing, (2). The bacterial infection is carried in the environment and transmitted by ticks which are the principal vectors of the bacterium. There can be several routes of infection in humans, including oral transmission via contaminated raw milk and dairy products although the inhalation of contaminated aerosols stands on top (3).
The infection can also be spread through the milk, fecal material, urine, vaginal secretions, especially at the time of abortion or parturition of the animals which are infected with C. burnetii (1). Q fever most commonly occurs in people who are working in the livestock industry including animal breeders, slaughterhouse workers, dairy industry staff, leather factory workers, or/and oil and industries that involve the transportation of the animals (4). This fever is often diagnosed only through a systematical consideration since its clinical manifestations are highly variable. In addition, only serology would provide the best approach to the distinction between acute and chronic infections. Further, Q fever in humans is usually manifested with no symptoms or as a mild flu-like disease which can recover spontaneously. However, some patients represent more serious states of the disease, some of which may lead to death. The risk factors of chronic disease include cardiac valve defects and vascular diseases, especially in immunocompromised hosts and pregnant women (3). According to Singh et al (5), the available methods for C. burnetii detection are as follows:
-
Classical methods, for example, culturing microorganisms in suitable cell lines;
-
Serological tests such as immunofluorescence, micro-immunofluorescence, complement fixation test, or the enzyme-linked immunosorbent assay (ELISA);
-
Molecular biology techniques like nested and real-time polymerase chain reaction (PCR).
Generally, dangerous and time-consuming Coxiella culturing is no longer performed for Q fever diagnosis since it is difficult. Hence, the use of diagnostic methods that are based on molecular biology techniques can significantly help the early diagnosis and treatment management of Q fever (6).
Objectives
No studies have so far investigated the role of cream and butter in C. burnetii infection with respect to Iran, and the rest of the world. Considering the low infectious dose of this pathogen and the lack of the availability of published data on the contamination of dairy products, the present study sought to evaluate the prevalence of C. burnetii in traditional and industrial butter and cream by nested PCR in Shahrekord, Iran.
Materials and Methods
Sampling
In total, 200 samples were collected from retailers located in different regions of Shahrekord, Iran during March–September 2016 that included: traditional cream samples (i.e., 10 goat, 20 sheep, and 39 bovine cream samples); traditional butter samples (i.e., 12 sheep and 69 bovine butter samples); industrial cream samples (25 bovine samples); and industrial butter samples (25 bovine samples). A cooler with ice packs was utilized to immediately transport the samples to a laboratory and process them in 1 hour after collection.
Using PCR to Definitely Diagnose Coxiella burnetii
Zhang et al and Fretz et al methods were utilized to detect C. burnetii in the samples (7,8). Then, 2 g of butter or 2 mL of cream was transferred to the tube by using a genomic DNA purification kit (Cinnagene, Iran), followed by performing the PCR with the purified extracted DNA. The quality and quantity of the extracted DNA were evaluated by spectrophotometry with a ratio of 260-280 nm wavelength (7,8).
The nested PCR was applied to detect the presence of the C. burnetii genomic DNA in the samples. In addition, the methods of Zhang et al and Fretz et al (7,8) was also employed for the sequences of the primers that were used for the com1 gene amplification (encoding the outer membrane of C. burnetii) (Table 1). Further, the optimum concentrations of substances with a final volume of 25 mL containing 5 μL of DNA sample, 0.5 mM MgCl2, 1 μM primer OMP1, 1 μM primer OMP2, 0.5 U/reaction of Taq DNA polymerase, and 0.2 mM (each) dNTPs (Fermetas, Germany) were used to perform the first stage of the PCR (9).
Table 1.
Primers for the Identification of C. burnetii
|
Primer
|
Sequence (5’ →3’)
|
Amplicon Size (bp)
|
| OMP1 |
5’-AGT AGA AGC ATC CCA AGC ATT G -’3 |
501 |
| OMP2 |
5’-TGC CTG CTA GCT GTA ACG ATT G -’3 |
| OMP3 |
5’-GAA GCG CAA CAA GAA GAA CAC- ‘3 |
438 |
| OMP4 |
5’TTG GAA GTT ATC ACG CAG TTG-’3 |
Furthermore, sterilized mineral oil (2–3 drops) was added to the PCR mixture in order to avoid contamination and evaporation. Next, the tubes were placed in an Eppendorf Master Cycler Gradient Thermal Cycler (Germany) and a temperature program was run for the PCR assay at 94°C for 4 minutes, and then for 30 cycles at 94°C for 1 minute, 56°C for 1 minute, and finally, at 72°C for 1 minute in a DNA thermal cycler (9).
For the second amplification, the reaction was performed with a total volume of 25 μL including 2 μL of the DNA sample, 0.5 mM of MgCl2, 0.2 mM (each) of dNTPs, 0.8 μM of OMP3, 0.8 μM of OMP4 primer, and 0.5 U/reaction of Taq DNA polymerase. The PCR assay was performed at 95°C for 4 minutes and then for 30 cycles at 94°C for 1 minute, 57°C for 1 minute, and 72°C for 1 minute. The PCR-amplified products (i.e., OMP1-OMP2: 501 bp & OMP3-OMP4: 438 bp) were examined by electrophoresis in a 1.5% agarose gel, stained with 1% solution of ethidium bromide, and then investigated under UV illumination. In the present study, the C. burnetii DNA (Serial Number: 3154; Genekam Biotechnology AG, Germany) and DNase− free water were used as positive and negative controls, respectively (9).
Statistical Analysis
Data were transferred to a Microsoft Excel spreadsheet (Microsoft Corp, Redmond, WA, USA) SPSS software, version 19.0 (SPSS Inc., Chicago, II, USA) and the chi-square test was applied to analyze the data. The differences were considered significant at P <0.05.
Results
A number of 200 traditional and industrial butter and cream samples were experimented in Shahrekord and were reported to have C. burnetiiusing the Nested PCR (Table 2 and Figure 1). In total, 4 out of 69 (5.79%) traditional bovine cream samples, 1 out of 20 (5%) traditional sheep butter samples, 1 out of 39 (2.56%) traditional bovine butter samples were positive for C. burnetii. The statistical analysis of the obtained results revealed no significant difference between the various samples and the prevalence of C. burnetii (P<0.05).
Table 2.
The Prevalence of Coxiella burnetii in the Samples of Traditional and Industrial Cream and Butter in Shahrekord
|
Samples
|
No. of Samples
|
No. (%) of
Coxiella
burnetii
Positive Samples
|
| Traditional cream |
Sheep |
12 |
0 (0) |
| Bovine |
69 |
4 (5.79) |
| Industrial cream |
Bovine |
25 |
0 (0) |
| Traditional butter |
Goat |
10 |
0 (0) |
| Sheep |
20 |
1 (5) |
| Bovine |
39 |
1 (2.56) |
| Industrial butter |
Bovine |
25 |
0 (0) |
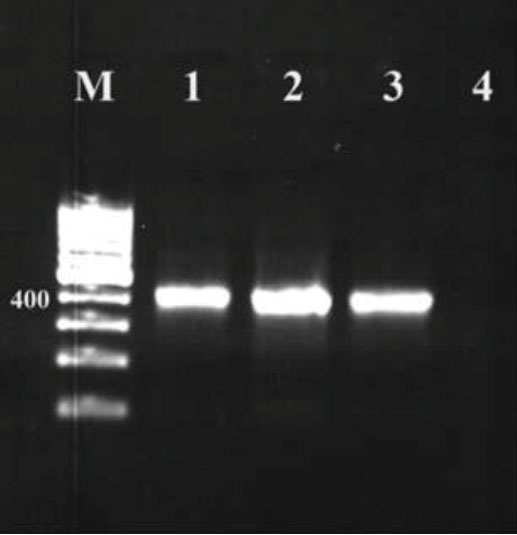
Figure 1.
Nested Polymerase Chain Reaction Coxiella burnetii Samples of Traditional and Industrial Cream and Butter in Shahrekord. Note. Column M: 100 bp DNA marker; Columns 1 and 2: Positive samples of C. burnetii; Column 3: Positive control; Column 4: Negative control.
.
Nested Polymerase Chain Reaction Coxiella burnetii Samples of Traditional and Industrial Cream and Butter in Shahrekord. Note. Column M: 100 bp DNA marker; Columns 1 and 2: Positive samples of C. burnetii; Column 3: Positive control; Column 4: Negative control.
Discussion
Overall, the results of this study demonstrated the highest and lowest rates of infection in the traditional bovine cream samples (4 out of 69 samples, 5.79%) and traditional bovine (1 out of 39, 2.56%) and sheep (1 out of 20, 5%) butter samples, respectively. Our findings could be compared with those of several other studies conducted in Iran and other countries.
For example, Muskens et al reported that 78.6% of the cow milk samples contained antibodies against C. burnetii and the prevalence of C. burnetii in the samples tested was found to be 56.6% using the PCR (10). The most important reasons for the difference in the prevalence of C. burnetii in cow milk samples across the different regions of the world are related to various types of geographic regions, the types of study methods, the type and the number of samples, the season of the year, as well as the sampling of infected and non-infected herds (11).
Similarly, Can et al used PCR with primers based on a repeat region, including transposon, for the direct detection of C. burnetiiin the milk samples. In their study, 150 milk samples (9%) were positive for C. burnetii, including 5 cow, 2 sheep, and 2 goat milk samples (12).
Other studies reported a different range of the presence of C. burnetiiin milk. About 1.8% of the total goat milk samples and 0% of sheep milk in Iran were tested positive (9) whereas, in Japan, only 17.1% of the cheese samples were reported to be positive (13). The occurrence of food-borne microbial diseases in both humans and animals is increasing in developing countries due to poor health. On the other hand, developed countries indicated almost no cases of infections or food poisoning due to their high standards for health. As a result, the determination of the detailed statistics of the strike rate, especially for the developing countries, is impossible. The food itself can act as a carrier of the infection. For instance, in cases where the infection was contracted from non-plant based food in which most of the infectious and non-infectious bodies, which (under some conditions) supported an infectious mass and acted either as an active or only a passive carrier, demonstrated that the infection could only be transmitted through food. The examples of such food items are milk and dairy products. Furthermore, nowadays, with the increased knowledge of the consumption of milk and dairy products, it is important to have healthy milk and dairy products, especially for the elderly and infants who have weaker immunity compared to other individuals (14). The available nutrients (e.g., calcium, protein, and vitamins A-D) in dairy products are essential for strong bones and teeth and they reduce the risk of osteoporosis, help maintain a healthy nervous system, activate muscle contractions, and promote skin and eye health. Although Q fever is primarily an occupational disease, the consumption of contaminated milk and dairy products can play an important role in the epidemiology of the disease in humans (14).
Rodolakis proposed a hypothesis that suggested that the consumption of dairy products from C. burnetii-contaminated animals can lead to its food-borne infection in humans (15). In the research by Huebner and Jellison, the pasteurization of dairy products could remove C. burnetii from contaminated milk (16). Based on the results of another study, the highest prevalence of serum and clinical diseases was observed in patients who consumed raw milk (17). However, the results of these studies showed that most of the people living in rural areas eat raw milk and are exposed to ruminating mammals, indicating possible contamination from aerosol inhalation. Nevertheless, an acute fever outbreak in Newfoundland, Canada, was found to occur by eating pasteurized goat cheese as an independent risk factor (18). Moreover, in a study conducted in a prison, C. burnetii serology revealed a higher risk in those consuming raw milk (19). Conversely, no repression or clinical illness was reported in the study by Krumbiegel and Wisniewski, which included 34 volunteers who drank raw milk (20). C. burnetii DNA was detected in 64% of the dairy products in France (17), especially in dairy cows with a higher percentage. Nonetheless, no live bacteria were removed from cheese and yogurt in the mentioned study. Consequently, the digestive pathway may play an important role in C. burnetii transmission even if it is not a major public health threat. The range of Q fever animal hosts is large and the disease spreads from ticks to primates. These microorganisms are found in the bodies of mollusks, arthropods, fish, birds, reptiles, marsupials, and livestock in many parts of the world (21).
Additionally, Nasehfar et al (22) found that clinically healthy dairy cows were the major sources of C. burnetii infection in Iran. In the current study, the milk samples of bovine bulk from central Iran were used to study the prevalence of C. burnetii. In the present study, 5 out of 100 (0.05%) bovine milk samples were tested positive for C. burnetii.
Q fever was first described by Derrick et al following the outbreak of flu-like illness among the slaughterhouse workers in Australia in 1937. It was considered as a zoonotic disease between livestock and humans, the spread of which was first reported in animals and then humans in Iran in 1952. The report of this disease in animals displayed evidence of a serological outbreak of fever while it was reported in humans in two patients showing severe fever and neurological symptoms associated with Q fever. In addition, this fever was first reported in Abadan, Khuzestan province, Iran. The unpasteurized samples of milk and dairy products in the study of Khanzadi et al (23) conducted in Mashhad, in Khorasan-Razavi province of Iran demonstrated positive touch-down PCR tests for C. burnetii in 2/28 cheese samples (7.14%), as well as 2/26 yoghurt samples (7.69%), 8/23 sheep milk samples (34.78%), and 2/60 bovine milk samples (3.33%).
Likewise, Kargaret al indicated that the prevalence of C. burnetii contamination in Jahrom in southern Iran was 11% after evaluating 100 bovine bulk milk samples (24).
In other cities such as Fars, Qom, Kerman, Khuzestan, and Yazd, Abbasi et al, using a nested PCR assay, reported 2% goat milk samples (6/296), 18.2% samples from the dairy herds of Fars (4/22), and 4.2% (1/24) samples from the product herds of Khuzestan. In addition, the results showed product herds of Yazd as 5.5% (1/18), the 6 positive samples for the presence of C. burnetii (25).
Coxiella burnetii can survive for at least 586 days in the dried feces of ticks, and 3 months on the moist soil corrals of birthing sheep (1). Various factors can cause a relatively high prevalence of coxiellosis in cows. In the study by Dhaka et al (26), its prevalence was discovered to be 29.91%. According to the researchers in this field, the contributing factors to coxiellosis prevalence were as follows:
-
Large herd size;
-
Frieswal herds since older animals are more likely to become infected after their first deliveries;
-
Inappropriate disposal of environmental wastes such as aborted materials and fertilizer on the farm;
-
Low bio-security measures that allow stray dogs and wild birds enter the farm;
-
High prevalence of reproductive disorders and other immune infections such as tuberculosis, brucellosis, Johne’s disease, and mastitis (26).
In ewes, C. burnetii secretion often occurs in the vagina and feces while secretion milk can be the source of most bacteria in cows and goats (27).
The results of this study and those of other studies indicated that traditional dairy products could be considered as an important reservoir of C. burnetii infection in Iran. C. burnetii is a dangerous bacterium with unique features resulting in the bio-safety rating of level 3. Hence, only experienced people are permitted to work with the infected samples of C. burnetii to study its culture and isolation in laboratories. Further, C. burnetii is an obligate intracellular bacterium whose culture needs a live host, which is time-consuming and costly and involves high risk. Despite the suitability of serological tests to recognize its presence, it takes several weeks to produce the antibodies to fight against C. burnetii. Fortunately, this problem is limited to the diagnosis of acute Q fever. All the mentioned factors become even more important for the early diagnosis of C. burnetii through the standard molecular biology techniques. Therefore, it is recommended to establish standard setups for the diagnostic molecular approaches which are suitable to be used in our country (1).
Conclusions
The results of this study showed that C. burnetiiwas absent in the industrial butter and cream samples and thus there was no danger to public health, thanks to the management methods of food safety systems such as Hazard Analysis and Critical Control Point, Good Manufacturing Practice, and Good Health Practice employed in the dairy factories in Iran. In this research, the results of the nested PCR tests on the traditional butter and cream samples revealed that the samples were contaminated with C. burnetii. The findings of this study and those of other studies conducted in Iran in recent years have shown that Q fever is present in Iran while it is not considered to be an important issue or is confused with other febrile illnesses such as brucellosis or influenza due to the existence of few available studies in this field. Given the nature of zoonosis and its possible contamination through a host animal, the veterinary authorities and ministry of health subsectors should necessarily participate in the extensive and proximity interactions to identify its reservoirs and prevent or control the disease. Regarding its health and economic importance in humans and livestock, it is suggested to identify the disease repository of ruminants after slaughter and to vaccinate those people at risk, including veterinarians and slaughterhouse workers.
Conflict of Interests
None.
Funding
This paper was extracted from project No. 94149 and was financially supported by the Young Researchers and Elite Club, Shahrekord Branch, Islamic Azad University, Shahrekord, Iran.
References
- Bauer AE, Olivas S, Cooper M, Hornstra H, Keim P, Pearson T. Estimated herd prevalence and sequence types of Coxiella burnetii in bulk tank milk samples from commercial dairies in Indiana. BMC Vet Res 2015; 11:186. doi: 10.1186/s12917-015-0517-3 [Crossref] [ Google Scholar]
- Angelakis E, Raoult D. Q fever. Vet Microbiol 2010; 140(3-4):297-309. doi: 10.1016/j.vetmic.2009.07.016 [Crossref] [ Google Scholar]
- Barlow J, Rauch B, Welcome F, Kim SG, Dubovi E, Schukken Y. Association between Coxiella burnetii shedding in milk and subclinical mastitis in dairy cattle. Vet Res 2008; 39(3):23. doi: 10.1051/vetres:2007060 [Crossref] [ Google Scholar]
- Kirkan F, Kaya O, Tekbiyik S, Parin U. Detection of Coxiella burnetii in cattle by PCR. Turk J Vet Anim Sci 2008; 32(3):215-20. [ Google Scholar]
- Singh J, Birbian N, Sinha S, Goswami A. A critical review on PCR, its types and applications. Int J Adv Res Biol Sci 2014; 1(7):65-80. [ Google Scholar]
- Tang YW, Stratton CW. Advanced Techniques in Diagnostic Microbiology. 2nd ed. Springer International Publishing; 2014.
- Zhang GQ, Nguyen SV, To H, Ogawa M, Hotta A, Yamaguchi T. Clinical evaluation of a new PCR assay for detection of Coxiella burnetii in human serum samples. J Clin Microbiol 1998; 36(1):77-80. [ Google Scholar]
- Fretz R, Schaeren W, Tanner M, Baumgartner A. Screening of various foodstuffs for occurrence of Coxiella burnetii in Switzerland. Int J Food Microbiol 2007; 116(3):414-8. doi: 10.1016/j.ijfoodmicro.2007.03.001 [Crossref] [ Google Scholar]
- Rahimi E. Coxiella burnetii in goat bulk milk samples in Iran. Afr J Microbiol Res 2010; 4(21):2324-6. [ Google Scholar]
- Muskens J, van Engelen E, van Maanen C, Bartels C, Lam TJ. Prevalence of Coxiella burnetii infection in Dutch dairy herds based on testing bulk tank milk and individual samples by PCR and ELISA. Vet Rec 2011; 168(3):79. doi: 10.1136/vr.c6106 [Crossref] [ Google Scholar]
- Rodolakis A, Berri M, Hechard C, Caudron C, Souriau A, Bodier CC. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J Dairy Sci 2007; 90(12):5352-60. doi: 10.3168/jds.2006-815 [Crossref] [ Google Scholar]
- Can HY, Elmalı M, Karagöz A. Detection of Coxiella burnetii in cows’, goats’, and ewes’ bulk milk samples using polymerase chain reaction (PCR). Mljekarstvo 2015; 65(1):26-31. doi: 10.15567/mljekarstvo.2015.0104 [Crossref] [ Google Scholar]
- Hirai A, Nakama A, Chiba T, Kai A. Development of a method for detecting Coxiella burnetii in cheese samples. J Vet Med Sci 2012; 74(2):175-80. doi: 10.1292/jvms.11-0023 [Crossref] [ Google Scholar]
- Park YW, Haenlein GFW. Milk and Dairy Products in Human Nutrition: Production, Composition and Health. 1st ed. Wiley-Blackwell; 2013. p. 1-24.
- Rodolakis A. Q Fever in dairy animals. Ann N Y Acad Sci 2009; 1166:90-3. doi: 10.1111/j.1749-6632.2009.04511.x [Crossref] [ Google Scholar]
- Huebner RJ, Jellison WL. Q fever studies in Southern California; effects of pasteurization on survival of C burneti in naturally infected milk. Public Health Rep 1949; 64(16):499-511. [ Google Scholar]
- Eldin C, Angelakis E, Renvoise A, Raoult D. Coxiella burnetii DNA, but not viable bacteria, in dairy products in France. Am J Trop Med Hyg 2013; 88(4):765-9. doi: 10.4269/ajtmh.12-0212 [Crossref] [ Google Scholar]
- Hatchette TF, Hudson RC, Schlech WF, Campbell NA, Hatchette JE, Ratnam S. Goat-associated Q fever: a new disease in Newfoundland. Emerg Infect Dis 2001; 7(3):413-9. doi: 10.3201/eid0703.010308 [Crossref] [ Google Scholar]
- Benson WW, Brock DW, Mather J. Serologic analysis of a penitentiary group using raw milk from a Q fever infected herd. Public Health Rep 1963; 78:707-10. [ Google Scholar]
- Krumbiegel ER, Wisniewski HJ. Q fever in the Milwaukee area II Consumption of infected raw milk by human volunteers. Arch Environ Health 1970; 21(1):63-5. doi: 10.1080/00039896.1970.10667193 [Crossref] [ Google Scholar]
- Marrie TJ, Raoult D. Coxiella burnetii (Q fever). 7th ed. In: Mandell, Douglas and bennett’s, editors. In: Mandell G, Bennett J, Dolin R, eds. Principles and practice of infecious diseases. Churchill Livingstone; 2009.
- Nasehfar A, Bonyadian M, Khadivi Boroujeni R, Mehmandoust Esfahani M, Kazemeini H, Momeni Shahraki M. Prevalence of Coxiella Burnetii by Nested PCR in Bovine Bulk Milk Samples in Central Zone of Iran. Am Adv J Biol Sci 2015; 1(1):10-3. [ Google Scholar]
- khanzadi s, Jamshidi A, Razmyar J, Borji S. Identification of Coxiella burnetii by touch-down PCR assay in unpasteurized milk and dairy products in North-East of Iran. Iranian Journal of Veterinary Medicine 2014; 8(1):15-9. doi: 10.22059/ijvm.2014.50558.[Persian] [Crossref] [ Google Scholar]
- Kargar M, Rashidi A, Doosti A, Ghorbani-Dalini S, Najafi A. Prevalence of Coxiella burnetii in bovine bulk milk samples in southern Iran. Comp Clin Path 2013; 22(3):331-4. doi: 10.1007/s00580-012-1406-9 [Crossref] [ Google Scholar]
- Abbasi S, Farzan R, Momtaz H. Molecular detection of Coxiella burnetii in goat bulk milk samples in some provinces of Iran. Afr J Biotechnol 2011; 10(80):18513-5. doi: 10.5897/AJB11.2508 [Crossref] [ Google Scholar]
- Dhaka P, Malik SS, Yadav JP, Kumar M, Baranwal A, Barbuddhe SB. Seroprevalence and molecular detection of coxiellosis among cattle and their human contacts in an organized dairy farm. J Infect Public Health 2019; 12(2):190-4. doi: 10.1016/j.jiph.2018.10.001 [Crossref] [ Google Scholar]
- Rodolakis A, Berri M, Héchard C, Caudron C, Souriau A, Bodier CC. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine, and ovine herds. J Dairy Sci 2007; 90(12):5352-60. doi: 10.3168/jds.2006-815 [Crossref] [ Google Scholar]